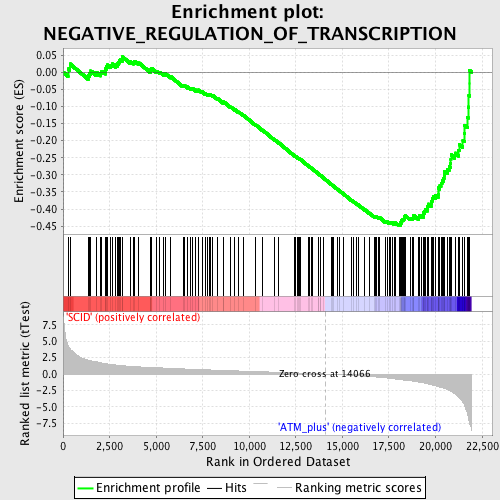
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NEGATIVE_REGULATION_OF_TRANSCRIPTION |
| Enrichment Score (ES) | -0.44929162 |
| Normalized Enrichment Score (NES) | -1.7347788 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.21891211 |
| FWER p-Value | 0.945 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DRAP1 | 1451676_at | 266 | 4.329 | 0.0096 | No | ||
| 2 | TIMELESS | 1417586_at 1417587_at | 369 | 3.824 | 0.0242 | No | ||
| 3 | HELLS | 1417541_at 1430139_at 1453361_at | 1345 | 2.097 | -0.0100 | No | ||
| 4 | NR0B1 | 1417760_at | 1425 | 2.039 | -0.0034 | No | ||
| 5 | GATAD2A | 1423992_at 1445239_at 1451197_s_at 1451198_at 1455505_at | 1493 | 2.004 | 0.0036 | No | ||
| 6 | MDM4 | 1419558_at 1453982_at 1460542_s_at | 1804 | 1.844 | -0.0013 | No | ||
| 7 | EPC1 | 1418850_at 1429914_at 1440198_at 1440742_at 1442279_at 1443675_at 1458315_at | 2019 | 1.711 | -0.0025 | No | ||
| 8 | PHB | 1435144_at 1442233_at 1448563_at | 2079 | 1.682 | 0.0032 | No | ||
| 9 | HDAC8 | 1428933_at | 2268 | 1.584 | 0.0026 | No | ||
| 10 | HEXIM2 | 1429025_a_at 1429026_at | 2286 | 1.576 | 0.0097 | No | ||
| 11 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 2317 | 1.556 | 0.0162 | No | ||
| 12 | GFI1B | 1420399_at | 2375 | 1.534 | 0.0213 | No | ||
| 13 | FOXD3 | 1422210_at | 2547 | 1.454 | 0.0207 | No | ||
| 14 | PHF21A | 1418390_at 1418391_at 1439123_at 1444679_at 1458166_at | 2628 | 1.427 | 0.0242 | No | ||
| 15 | KLF11 | 1437241_at | 2839 | 1.355 | 0.0214 | No | ||
| 16 | BRCA1 | 1424629_at 1424630_a_at 1427698_at 1443763_at 1451417_at | 2922 | 1.334 | 0.0244 | No | ||
| 17 | VPS72 | 1418284_at | 2980 | 1.317 | 0.0284 | No | ||
| 18 | E2F6 | 1437914_at 1448835_at | 3016 | 1.307 | 0.0334 | No | ||
| 19 | PIAS4 | 1418861_at 1455394_at | 3080 | 1.289 | 0.0370 | No | ||
| 20 | LANCL2 | 1420856_a_at 1420857_at 1450028_a_at | 3181 | 1.260 | 0.0387 | No | ||
| 21 | TNP1 | 1415924_at 1438632_x_at | 3187 | 1.258 | 0.0448 | No | ||
| 22 | DMBX1 | 1460277_at | 3629 | 1.159 | 0.0304 | No | ||
| 23 | GLIS1 | 1425423_at | 3774 | 1.135 | 0.0295 | No | ||
| 24 | ID1 | 1425895_a_at | 3846 | 1.120 | 0.0319 | No | ||
| 25 | PRDM1 | 1420425_at 1444997_at | 4071 | 1.083 | 0.0271 | No | ||
| 26 | WWP1 | 1427097_at 1427098_at 1446364_at 1446853_at 1452299_at | 4708 | 1.000 | 0.0029 | No | ||
| 27 | ID4 | 1423259_at 1423260_at 1438441_at 1450928_at | 4709 | 1.000 | 0.0079 | No | ||
| 28 | HMGB1 | 1416176_at 1425048_a_at 1435324_x_at | 4761 | 0.994 | 0.0106 | No | ||
| 29 | GLIS2 | 1416598_at 1416599_at | 5025 | 0.962 | 0.0034 | No | ||
| 30 | MECP2 | 1438930_s_at 1449266_at 1460246_at | 5194 | 0.938 | 0.0004 | No | ||
| 31 | NSD1 | 1420881_at 1435088_at 1440972_at 1456916_at | 5394 | 0.909 | -0.0042 | No | ||
| 32 | RYBP | 1421111_at | 5493 | 0.896 | -0.0042 | No | ||
| 33 | SNAI2 | 1418673_at 1447643_x_at | 5774 | 0.858 | -0.0127 | No | ||
| 34 | TCEAL1 | 1424634_at | 6438 | 0.776 | -0.0392 | No | ||
| 35 | EREG | 1419431_at | 6521 | 0.765 | -0.0391 | No | ||
| 36 | TWIST1 | 1418733_at | 6685 | 0.746 | -0.0429 | No | ||
| 37 | ERN2 | 1450139_at | 6844 | 0.729 | -0.0464 | No | ||
| 38 | ENO1 | 1419023_x_at | 6937 | 0.717 | -0.0471 | No | ||
| 39 | TRIM27 | 1415815_at 1438376_s_at 1441051_at 1448101_s_at 1456375_x_at 1457051_at | 7117 | 0.697 | -0.0518 | No | ||
| 40 | TLE1 | 1422751_at 1434033_at | 7248 | 0.683 | -0.0543 | No | ||
| 41 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 7270 | 0.679 | -0.0518 | No | ||
| 42 | PPARD | 1425703_at 1439797_at | 7462 | 0.655 | -0.0573 | No | ||
| 43 | RUNX2 | 1424704_at 1425389_a_at 1426034_a_at | 7633 | 0.638 | -0.0619 | No | ||
| 44 | SIRT1 | 1418640_at 1458538_at | 7772 | 0.623 | -0.0651 | No | ||
| 45 | NKX2-5 | 1449566_at | 7848 | 0.616 | -0.0654 | No | ||
| 46 | NR1I2 | 1425723_at 1451807_at | 7903 | 0.610 | -0.0649 | No | ||
| 47 | ILF3 | 1422546_at 1436802_at 1460669_at | 8047 | 0.595 | -0.0684 | No | ||
| 48 | PAWR | 1444808_at 1445390_at | 8317 | 0.568 | -0.0779 | No | ||
| 49 | GLI2 | 1446086_s_at 1459211_at | 8601 | 0.539 | -0.0882 | No | ||
| 50 | HSBP1 | 1423906_at 1451162_at | 8627 | 0.537 | -0.0866 | No | ||
| 51 | NDUFA13 | 1430713_s_at | 9007 | 0.499 | -0.1015 | No | ||
| 52 | TNF | 1419607_at | 9219 | 0.478 | -0.1088 | No | ||
| 53 | PCGF6 | 1424081_at 1432108_at 1454120_a_at | 9431 | 0.458 | -0.1162 | No | ||
| 54 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 9676 | 0.432 | -0.1252 | No | ||
| 55 | BMP2 | 1423635_at | 10353 | 0.369 | -0.1544 | No | ||
| 56 | JARID1B | 1427142_s_at 1427143_at 1442278_at 1457482_at | 10706 | 0.335 | -0.1688 | No | ||
| 57 | ATP8B1 | 1420075_at 1420076_at 1455396_at | 11367 | 0.273 | -0.1978 | No | ||
| 58 | CALCA | 1427355_at 1452004_at | 11578 | 0.251 | -0.2061 | No | ||
| 59 | RSF1 | 1438735_at | 12431 | 0.170 | -0.2444 | No | ||
| 60 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 12497 | 0.165 | -0.2465 | No | ||
| 61 | RBM9 | 1418245_a_at 1418246_at 1418247_s_at 1434938_at | 12607 | 0.155 | -0.2508 | No | ||
| 62 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 12643 | 0.151 | -0.2516 | No | ||
| 63 | ID2 | 1422537_a_at 1435176_a_at 1453596_at | 12717 | 0.145 | -0.2542 | No | ||
| 64 | NR0B2 | 1449854_at | 12761 | 0.141 | -0.2555 | No | ||
| 65 | NR6A1 | 1421515_at 1421516_at 1428825_at 1428826_at 1445872_at | 13153 | 0.100 | -0.2729 | No | ||
| 66 | GRLF1 | 1434009_at 1440675_at 1442181_at | 13232 | 0.089 | -0.2761 | No | ||
| 67 | FOSB | 1422134_at | 13344 | 0.077 | -0.2808 | No | ||
| 68 | MYST4 | 1423508_at 1446189_at 1447258_at 1447758_x_at | 13408 | 0.071 | -0.2833 | No | ||
| 69 | FST | 1421365_at 1434458_at | 13729 | 0.036 | -0.2978 | No | ||
| 70 | ZBTB32 | 1432459_a_at 1447330_at | 13735 | 0.035 | -0.2979 | No | ||
| 71 | SIRT5 | 1428915_at 1428916_s_at 1432105_at | 13811 | 0.026 | -0.3012 | No | ||
| 72 | MYST1 | 1432080_s_at | 13831 | 0.024 | -0.3019 | No | ||
| 73 | KLF12 | 1439846_at 1439847_s_at 1441040_at 1455521_at | 13967 | 0.010 | -0.3081 | No | ||
| 74 | IRF2 | 1418265_s_at 1447527_at | 14417 | -0.043 | -0.3285 | No | ||
| 75 | TCF25 | 1424053_a_at 1437204_a_at 1437205_at 1451081_a_at 1459240_at 1460109_at | 14469 | -0.050 | -0.3306 | No | ||
| 76 | MDFI | 1420713_a_at 1425924_at | 14527 | -0.057 | -0.3329 | No | ||
| 77 | BCLAF1 | 1428844_a_at 1428845_at 1436023_at 1438089_a_at | 14729 | -0.083 | -0.3417 | No | ||
| 78 | RFX3 | 1425413_at 1441253_at 1441702_at | 14846 | -0.097 | -0.3466 | No | ||
| 79 | TWIST2 | 1448925_at | 15046 | -0.124 | -0.3551 | No | ||
| 80 | FOXP3 | 1420765_a_at 1457082_at | 15507 | -0.190 | -0.3752 | No | ||
| 81 | DEDD | 1432000_a_at 1434994_at 1434995_s_at | 15595 | -0.204 | -0.3782 | No | ||
| 82 | ZNF24 | 1422810_at 1426895_at 1426896_at 1430650_at 1430651_s_at 1450762_s_at | 15782 | -0.231 | -0.3856 | No | ||
| 83 | SIRT4 | 1426847_at | 15851 | -0.243 | -0.3875 | No | ||
| 84 | GLIS3 | 1430353_at 1443157_at 1443253_at | 16208 | -0.311 | -0.4022 | No | ||
| 85 | ATXN1 | 1438294_at 1445695_at 1450499_at 1457605_at | 16447 | -0.358 | -0.4114 | No | ||
| 86 | YAF2 | 1423399_a_at 1429839_a_at 1446073_at | 16738 | -0.430 | -0.4225 | No | ||
| 87 | NRG1 | 1456524_at | 16777 | -0.440 | -0.4220 | No | ||
| 88 | NKRF | 1434398_at 1441325_at 1445983_at | 16829 | -0.451 | -0.4221 | No | ||
| 89 | ING4 | 1448054_at 1460728_s_at | 16951 | -0.485 | -0.4252 | No | ||
| 90 | TBX3 | 1437479_x_at 1439567_at 1444594_at 1445757_at 1448029_at | 16977 | -0.491 | -0.4239 | No | ||
| 91 | KLF4 | 1417394_at 1417395_at | 17293 | -0.578 | -0.4354 | No | ||
| 92 | PEX14 | 1419053_at 1458153_at | 17397 | -0.606 | -0.4371 | No | ||
| 93 | NR1H2 | 1416353_at | 17509 | -0.645 | -0.4390 | No | ||
| 94 | PML | 1448757_at 1456103_at 1459137_at | 17575 | -0.667 | -0.4386 | No | ||
| 95 | CENPF | 1427161_at 1452334_at 1458447_at | 17679 | -0.695 | -0.4398 | No | ||
| 96 | DAXX | 1419026_at | 17806 | -0.738 | -0.4419 | No | ||
| 97 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 17852 | -0.755 | -0.4402 | No | ||
| 98 | ORC2L | 1418225_at 1418226_at 1418227_at | 18052 | -0.830 | -0.4451 | Yes | ||
| 99 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 18107 | -0.846 | -0.4433 | Yes | ||
| 100 | LDB1 | 1452024_a_at | 18109 | -0.846 | -0.4391 | Yes | ||
| 101 | SPI1 | 1418747_at | 18153 | -0.865 | -0.4367 | Yes | ||
| 102 | POU4F2 | 1437588_at | 18173 | -0.869 | -0.4332 | Yes | ||
| 103 | VDR | 1418175_at 1418176_at | 18209 | -0.885 | -0.4304 | Yes | ||
| 104 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 18298 | -0.913 | -0.4298 | Yes | ||
| 105 | PHF12 | 1434922_at 1453579_at | 18315 | -0.918 | -0.4259 | Yes | ||
| 106 | SNX6 | 1425148_a_at 1429497_s_at 1447234_s_at 1451602_at | 18324 | -0.920 | -0.4217 | Yes | ||
| 107 | MYST3 | 1436315_at 1437487_at 1446143_at 1446405_at | 18378 | -0.937 | -0.4194 | Yes | ||
| 108 | ARID5B | 1420972_at 1420973_at 1434283_at 1442176_at 1445423_at 1456831_at 1456973_at 1457483_at 1458238_at | 18637 | -1.019 | -0.4261 | Yes | ||
| 109 | SLA2 | 1437504_at 1451492_at | 18752 | -1.059 | -0.4260 | Yes | ||
| 110 | ZHX3 | 1419791_at 1441454_at 1442963_at 1452977_at 1454289_at 1456979_at 1457710_at | 18814 | -1.083 | -0.4234 | Yes | ||
| 111 | ARID5A | 1451340_at | 18817 | -1.084 | -0.4180 | Yes | ||
| 112 | SMAD2 | 1420634_a_at 1458304_at | 19111 | -1.219 | -0.4253 | Yes | ||
| 113 | GCLC | 1424296_at | 19122 | -1.228 | -0.4196 | Yes | ||
| 114 | PER1 | 1449851_at | 19247 | -1.290 | -0.4188 | Yes | ||
| 115 | NRIP1 | 1418469_at 1432603_at 1434384_at 1446389_at 1447211_at 1449089_at 1454295_at | 19351 | -1.349 | -0.4167 | Yes | ||
| 116 | MXD4 | 1422574_at 1434378_a_at 1434379_at | 19366 | -1.356 | -0.4105 | Yes | ||
| 117 | KLF10 | 1416029_at | 19419 | -1.390 | -0.4059 | Yes | ||
| 118 | SUPT5H | 1424255_at | 19444 | -1.406 | -0.4000 | Yes | ||
| 119 | PDCD4 | 1444171_at | 19569 | -1.485 | -0.3982 | Yes | ||
| 120 | BCOR | 1428773_s_at 1429438_at 1452910_at | 19579 | -1.490 | -0.3911 | Yes | ||
| 121 | ZNF148 | 1418381_at 1427730_a_at 1436217_at 1441612_at 1441701_at 1449068_at 1449069_at 1455483_at | 19615 | -1.511 | -0.3851 | Yes | ||
| 122 | SMARCE1 | 1422675_at 1422676_at 1430822_at | 19764 | -1.620 | -0.3837 | Yes | ||
| 123 | SIRT2 | 1423507_a_at | 19790 | -1.641 | -0.3766 | Yes | ||
| 124 | SNW1 | 1429002_at 1429003_at 1459026_at | 19817 | -1.661 | -0.3694 | Yes | ||
| 125 | SUMO1 | 1438289_a_at 1444557_at 1451005_at 1456349_x_at | 19865 | -1.693 | -0.3631 | Yes | ||
| 126 | ZHX1 | 1448875_at 1458382_a_at | 20011 | -1.813 | -0.3606 | Yes | ||
| 127 | SIN3A | 1419101_at 1419102_at 1419900_at 1449343_s_at | 20139 | -1.923 | -0.3568 | Yes | ||
| 128 | DR1 | 1416018_at 1416019_at | 20155 | -1.936 | -0.3477 | Yes | ||
| 129 | ELK3 | 1417662_at 1442986_at 1445886_at 1448797_at | 20156 | -1.938 | -0.3379 | Yes | ||
| 130 | IRF8 | 1416714_at 1448452_at | 20238 | -1.993 | -0.3316 | Yes | ||
| 131 | MEN1 | 1416347_at 1416348_at 1443826_x_at | 20296 | -2.021 | -0.3241 | Yes | ||
| 132 | KHDRBS1 | 1418628_at 1418630_at | 20352 | -2.058 | -0.3162 | Yes | ||
| 133 | HEXIM1 | 1419359_at 1425937_a_at | 20443 | -2.144 | -0.3096 | Yes | ||
| 134 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 20461 | -2.164 | -0.2995 | Yes | ||
| 135 | DNMT1 | 1422946_a_at 1435122_x_at 1445637_at 1447877_x_at | 20497 | -2.200 | -0.2900 | Yes | ||
| 136 | CTCF | 1418330_at 1449042_at | 20645 | -2.384 | -0.2847 | Yes | ||
| 137 | ZFP161 | 1420864_at 1420865_at 1420866_at 1437585_x_at | 20730 | -2.488 | -0.2761 | Yes | ||
| 138 | CSDA | 1435800_a_at 1451012_a_at | 20792 | -2.563 | -0.2660 | Yes | ||
| 139 | ZBTB16 | 1419874_x_at 1427638_at 1439163_at 1445562_at 1456986_at 1459557_at | 20826 | -2.618 | -0.2543 | Yes | ||
| 140 | SMARCC2 | 1428382_at | 20832 | -2.626 | -0.2413 | Yes | ||
| 141 | UBP1 | 1455222_a_at 1460655_a_at | 21046 | -2.985 | -0.2361 | Yes | ||
| 142 | IRF7 | 1417244_a_at | 21245 | -3.481 | -0.2276 | Yes | ||
| 143 | ZHX2 | 1455210_at | 21272 | -3.557 | -0.2109 | Yes | ||
| 144 | SMAD4 | 1422485_at 1422486_a_at 1422487_at 1444205_at | 21473 | -4.259 | -0.1987 | Yes | ||
| 145 | HDAC5 | 1415743_at | 21541 | -4.626 | -0.1784 | Yes | ||
| 146 | BCL3 | 1418133_at | 21555 | -4.716 | -0.1553 | Yes | ||
| 147 | TGFB1 | 1420653_at 1445360_at | 21716 | -6.029 | -0.1323 | Yes | ||
| 148 | ARID4A | 1436191_at 1438464_at 1456420_at | 21755 | -6.438 | -0.1016 | Yes | ||
| 149 | ZMYND11 | 1426531_at 1426532_at 1436153_a_at 1444461_at 1446182_at | 21792 | -6.999 | -0.0681 | Yes | ||
| 150 | ID3 | 1416630_at | 21832 | -7.406 | -0.0326 | Yes | ||
| 151 | BCL6 | 1421818_at 1450381_a_at | 21833 | -7.407 | 0.0047 | Yes |

