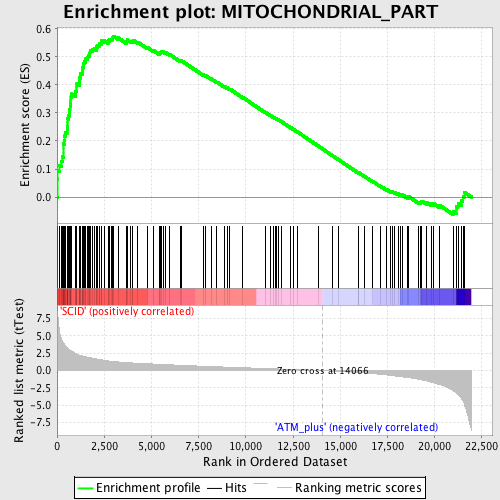
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | 0.57386637 |
| Normalized Enrichment Score (NES) | 2.2582877 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MRPS22 | 1416595_at 1443032_at | 18 | 7.986 | 0.0319 | Yes | ||
| 2 | TIMM50 | 1441819_x_at 1455456_a_at | 19 | 7.985 | 0.0646 | Yes | ||
| 3 | NFS1 | 1416373_at 1431431_a_at 1442017_at | 28 | 7.814 | 0.0962 | Yes | ||
| 4 | MRPS10 | 1428153_at 1458999_at | 145 | 5.327 | 0.1127 | Yes | ||
| 5 | NDUFS7 | 1424313_a_at 1451312_at | 219 | 4.633 | 0.1283 | Yes | ||
| 6 | ATP5G1 | 1416020_a_at 1444874_at | 272 | 4.304 | 0.1435 | Yes | ||
| 7 | TIMM10 | 1417499_at | 323 | 3.974 | 0.1575 | Yes | ||
| 8 | SDHD | 1428235_at 1437489_x_at | 324 | 3.973 | 0.1738 | Yes | ||
| 9 | ATP5G2 | 1415980_at 1456128_at | 325 | 3.967 | 0.1900 | Yes | ||
| 10 | ABCB6 | 1422524_at | 378 | 3.793 | 0.2032 | Yes | ||
| 11 | CS | 1422577_at 1422578_at 1450667_a_at | 390 | 3.745 | 0.2180 | Yes | ||
| 12 | ATP5E | 1416567_s_at | 446 | 3.593 | 0.2302 | Yes | ||
| 13 | MRPL55 | 1429453_a_at | 528 | 3.351 | 0.2402 | Yes | ||
| 14 | MRPS15 | 1430100_at 1449294_at 1456109_a_at | 538 | 3.329 | 0.2534 | Yes | ||
| 15 | MRPL23 | 1416948_at | 549 | 3.290 | 0.2664 | Yes | ||
| 16 | VDAC3 | 1416175_a_at | 551 | 3.284 | 0.2798 | Yes | ||
| 17 | ETFB | 1428181_at | 615 | 3.153 | 0.2898 | Yes | ||
| 18 | MRPL12 | 1452048_at | 642 | 3.075 | 0.3012 | Yes | ||
| 19 | MRPS36 | 1423242_at | 656 | 3.044 | 0.3131 | Yes | ||
| 20 | NDUFA9 | 1416663_at | 683 | 2.972 | 0.3241 | Yes | ||
| 21 | NDUFV1 | 1415966_a_at 1415967_at 1456015_x_at | 723 | 2.891 | 0.3341 | Yes | ||
| 22 | MRPS35 | 1452111_at | 731 | 2.875 | 0.3456 | Yes | ||
| 23 | MRPS28 | 1452585_at | 734 | 2.871 | 0.3572 | Yes | ||
| 24 | BCS1L | 1451541_at | 759 | 2.821 | 0.3677 | Yes | ||
| 25 | MRPS16 | 1448869_a_at | 969 | 2.487 | 0.3683 | Yes | ||
| 26 | UQCRC1 | 1428782_a_at | 988 | 2.464 | 0.3776 | Yes | ||
| 27 | BAX | 1416837_at | 1046 | 2.396 | 0.3848 | Yes | ||
| 28 | NDUFB6 | 1434056_a_at 1434057_at | 1047 | 2.394 | 0.3946 | Yes | ||
| 29 | TIMM13 | 1455211_a_at | 1050 | 2.391 | 0.4043 | Yes | ||
| 30 | MRPS18C | 1427901_at | 1171 | 2.233 | 0.4079 | Yes | ||
| 31 | ATP5O | 1416278_a_at | 1186 | 2.221 | 0.4163 | Yes | ||
| 32 | ETFA | 1423972_at | 1191 | 2.216 | 0.4252 | Yes | ||
| 33 | SUPV3L1 | 1432623_at 1460557_at | 1218 | 2.194 | 0.4330 | Yes | ||
| 34 | PITRM1 | 1426453_at 1456577_x_at | 1253 | 2.167 | 0.4403 | Yes | ||
| 35 | ABCB8 | 1422015_a_at 1423713_at 1460022_at | 1357 | 2.088 | 0.4442 | Yes | ||
| 36 | CYCS | 1422484_at 1445484_at | 1361 | 2.084 | 0.4526 | Yes | ||
| 37 | ACN9 | 1456614_at | 1364 | 2.081 | 0.4610 | Yes | ||
| 38 | NDUFS8 | 1423907_a_at 1423908_at 1434212_at 1434213_x_at 1434579_x_at 1455283_x_at | 1384 | 2.070 | 0.4686 | Yes | ||
| 39 | MRPL40 | 1448849_at | 1386 | 2.068 | 0.4770 | Yes | ||
| 40 | ALDH4A1 | 1452375_at | 1431 | 2.035 | 0.4833 | Yes | ||
| 41 | SLC25A1 | 1428190_at | 1501 | 2.001 | 0.4884 | Yes | ||
| 42 | ATP5F1 | 1426742_at 1433562_s_at | 1524 | 1.992 | 0.4955 | Yes | ||
| 43 | TIMM23 | 1416485_at | 1627 | 1.937 | 0.4988 | Yes | ||
| 44 | GRPEL1 | 1417320_at | 1638 | 1.933 | 0.5062 | Yes | ||
| 45 | VDAC1 | 1415998_at 1436992_x_at 1437192_x_at 1437452_x_at 1437947_x_at | 1719 | 1.891 | 0.5103 | Yes | ||
| 46 | COX6B2 | 1435275_at | 1740 | 1.878 | 0.5171 | Yes | ||
| 47 | MRPS12 | 1448799_s_at | 1780 | 1.854 | 0.5229 | Yes | ||
| 48 | NDUFA6 | 1448427_at | 1879 | 1.795 | 0.5257 | Yes | ||
| 49 | TOMM22 | 1429103_at 1434420_x_at | 1967 | 1.747 | 0.5289 | Yes | ||
| 50 | PHB | 1435144_at 1442233_at 1448563_at | 2079 | 1.682 | 0.5307 | Yes | ||
| 51 | SLC25A3 | 1416300_a_at 1440302_at | 2088 | 1.675 | 0.5372 | Yes | ||
| 52 | ATP5J | 1416143_at | 2155 | 1.642 | 0.5409 | Yes | ||
| 53 | NDUFS2 | 1451096_at | 2223 | 1.610 | 0.5444 | Yes | ||
| 54 | MRPS21 | 1422451_at | 2265 | 1.587 | 0.5490 | Yes | ||
| 55 | SLC25A15 | 1420966_at 1420967_at | 2343 | 1.547 | 0.5518 | Yes | ||
| 56 | ATP5B | 1416829_at | 2361 | 1.539 | 0.5573 | Yes | ||
| 57 | POLG2 | 1450816_at | 2489 | 1.480 | 0.5576 | Yes | ||
| 58 | NDUFA2 | 1417368_s_at | 2704 | 1.402 | 0.5535 | Yes | ||
| 59 | MPV17 | 1420387_at | 2723 | 1.395 | 0.5584 | Yes | ||
| 60 | MTX2 | 1430500_s_at | 2772 | 1.381 | 0.5618 | Yes | ||
| 61 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 2886 | 1.346 | 0.5622 | Yes | ||
| 62 | NDUFS3 | 1423737_at | 2935 | 1.330 | 0.5654 | Yes | ||
| 63 | NDUFA1 | 1422241_a_at | 2955 | 1.324 | 0.5699 | Yes | ||
| 64 | TIMM9 | 1428010_at 1449886_a_at | 2988 | 1.315 | 0.5739 | Yes | ||
| 65 | ABCF2 | 1423863_at | 3240 | 1.245 | 0.5675 | No | ||
| 66 | TIMM17A | 1426256_at | 3657 | 1.154 | 0.5531 | No | ||
| 67 | TIMM8B | 1431665_a_at | 3694 | 1.148 | 0.5561 | No | ||
| 68 | VDAC2 | 1415990_at | 3705 | 1.147 | 0.5604 | No | ||
| 69 | HSD3B2 | 1425127_at | 3909 | 1.112 | 0.5556 | No | ||
| 70 | MAOB | 1434354_at | 4008 | 1.091 | 0.5556 | No | ||
| 71 | MSTO1 | 1424577_at | 4017 | 1.090 | 0.5597 | No | ||
| 72 | ATP5D | 1423716_s_at 1456580_s_at | 4261 | 1.052 | 0.5529 | No | ||
| 73 | ATP5C1 | 1416058_s_at 1438809_at | 4775 | 0.992 | 0.5334 | No | ||
| 74 | PHB2 | 1416202_at 1438938_x_at 1455713_x_at 1460035_at | 5095 | 0.952 | 0.5227 | No | ||
| 75 | UCP3 | 1420657_at 1420658_at | 5442 | 0.903 | 0.5105 | No | ||
| 76 | MRPS24 | 1438563_s_at 1450865_s_at | 5448 | 0.902 | 0.5140 | No | ||
| 77 | MRPL10 | 1418112_at | 5467 | 0.900 | 0.5168 | No | ||
| 78 | NDUFS1 | 1425143_a_at 1453354_at | 5524 | 0.892 | 0.5179 | No | ||
| 79 | MFN2 | 1441424_at 1448131_at 1451900_at | 5608 | 0.882 | 0.5177 | No | ||
| 80 | NDUFS4 | 1418117_at 1438166_x_at 1444464_at 1448959_at 1459760_at | 5725 | 0.865 | 0.5159 | No | ||
| 81 | BCKDK | 1443813_x_at 1460644_at | 5937 | 0.838 | 0.5097 | No | ||
| 82 | TIMM44 | 1417670_at 1439371_x_at 1448801_a_at | 6523 | 0.765 | 0.4860 | No | ||
| 83 | HSD3B1 | 1448453_at | 6565 | 0.760 | 0.4872 | No | ||
| 84 | ATP5G3 | 1454661_at | 7732 | 0.627 | 0.4363 | No | ||
| 85 | FIS1 | 1416764_at 1443819_x_at | 7866 | 0.614 | 0.4327 | No | ||
| 86 | DNAJA3 | 1420629_a_at 1432066_at 1449935_a_at | 8161 | 0.584 | 0.4216 | No | ||
| 87 | TIMM17B | 1438656_x_at 1460685_at | 8465 | 0.553 | 0.4100 | No | ||
| 88 | CENTA2 | 1425638_at | 8857 | 0.515 | 0.3942 | No | ||
| 89 | NDUFA13 | 1430713_s_at | 9007 | 0.499 | 0.3894 | No | ||
| 90 | TFB2M | 1423441_at | 9155 | 0.484 | 0.3846 | No | ||
| 91 | SURF1 | 1450561_a_at | 9843 | 0.417 | 0.3548 | No | ||
| 92 | BNIP3 | 1422470_at | 11032 | 0.306 | 0.3016 | No | ||
| 93 | OXA1L | 1423738_at 1451088_a_at 1456845_at | 11302 | 0.281 | 0.2904 | No | ||
| 94 | MRPL52 | 1415761_at 1415762_x_at 1455366_at 1460701_a_at | 11468 | 0.264 | 0.2839 | No | ||
| 95 | NR3C1 | 1421866_at 1421867_at 1453860_s_at 1457635_s_at 1460303_at | 11557 | 0.254 | 0.2809 | No | ||
| 96 | BCKDHB | 1427153_at | 11617 | 0.248 | 0.2792 | No | ||
| 97 | MRPS11 | 1455233_at 1460195_at | 11702 | 0.239 | 0.2764 | No | ||
| 98 | MRPL32 | 1424372_at | 11889 | 0.218 | 0.2687 | No | ||
| 99 | UQCRH | 1452133_at 1453229_s_at | 12385 | 0.175 | 0.2467 | No | ||
| 100 | PMPCA | 1424661_at 1424662_at | 12499 | 0.165 | 0.2422 | No | ||
| 101 | SDHA | 1426688_at 1426689_s_at 1445317_at | 12709 | 0.145 | 0.2332 | No | ||
| 102 | CASQ1 | 1422598_at | 13853 | 0.023 | 0.1809 | No | ||
| 103 | MRPL41 | 1428588_a_at 1428589_at 1428590_at | 14561 | -0.060 | 0.1487 | No | ||
| 104 | SLC25A22 | 1452653_at | 14904 | -0.104 | 0.1335 | No | ||
| 105 | RAF1 | 1416078_s_at 1425419_a_at | 15969 | -0.268 | 0.0858 | No | ||
| 106 | IMMT | 1429533_at 1429534_a_at 1433470_a_at 1437126_at | 15970 | -0.268 | 0.0869 | No | ||
| 107 | GATM | 1423569_at | 16274 | -0.325 | 0.0743 | No | ||
| 108 | BCKDHA | 1416647_at | 16697 | -0.419 | 0.0567 | No | ||
| 109 | HTRA2 | 1436269_s_at 1460495_s_at | 17121 | -0.526 | 0.0394 | No | ||
| 110 | COX15 | 1426693_x_at 1437982_x_at 1452146_a_at 1460376_a_at | 17448 | -0.622 | 0.0270 | No | ||
| 111 | HADHB | 1437172_x_at | 17671 | -0.693 | 0.0197 | No | ||
| 112 | RAB11FIP5 | 1427405_s_at 1434314_s_at | 17753 | -0.717 | 0.0189 | No | ||
| 113 | PPOX | 1416618_at 1441862_at 1448065_at | 17894 | -0.769 | 0.0156 | No | ||
| 114 | ABCB7 | 1419931_at 1427490_at 1435006_s_at | 18058 | -0.832 | 0.0116 | No | ||
| 115 | OGDH | 1445632_at 1451274_at | 18163 | -0.867 | 0.0103 | No | ||
| 116 | UQCRB | 1416337_at 1455997_a_at | 18297 | -0.912 | 0.0080 | No | ||
| 117 | RHOT1 | 1423533_a_at 1440853_at | 18559 | -0.996 | 0.0001 | No | ||
| 118 | MRPL51 | 1416877_a_at 1416878_at 1416879_at | 18605 | -1.008 | 0.0022 | No | ||
| 119 | MRPS18A | 1416984_at 1457813_at | 19155 | -1.245 | -0.0179 | No | ||
| 120 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 19271 | -1.302 | -0.0179 | No | ||
| 121 | NNT | 1416105_at 1432608_at 1456573_x_at | 19316 | -1.330 | -0.0144 | No | ||
| 122 | HCCS | 1420889_at 1420890_at | 19568 | -1.484 | -0.0199 | No | ||
| 123 | MCL1 | 1416880_at 1416881_at 1437527_x_at 1448503_at 1456243_x_at 1456381_x_at | 19804 | -1.648 | -0.0239 | No | ||
| 124 | TOMM34 | 1448345_at | 19916 | -1.733 | -0.0219 | No | ||
| 125 | OPA1 | 1418768_at 1434890_at 1449214_a_at | 20236 | -1.993 | -0.0284 | No | ||
| 126 | RHOT2 | 1426822_at | 20972 | -2.853 | -0.0504 | No | ||
| 127 | DBT | 1440016_at 1449118_at | 21157 | -3.247 | -0.0455 | No | ||
| 128 | SLC25A11 | 1426586_at | 21160 | -3.255 | -0.0323 | No | ||
| 129 | CASP7 | 1426062_a_at 1448659_at | 21233 | -3.447 | -0.0215 | No | ||
| 130 | ALAS2 | 1451675_a_at | 21415 | -4.037 | -0.0133 | No | ||
| 131 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 21497 | -4.358 | 0.0009 | No | ||
| 132 | ACADM | 1415984_at | 21553 | -4.696 | 0.0176 | No |

