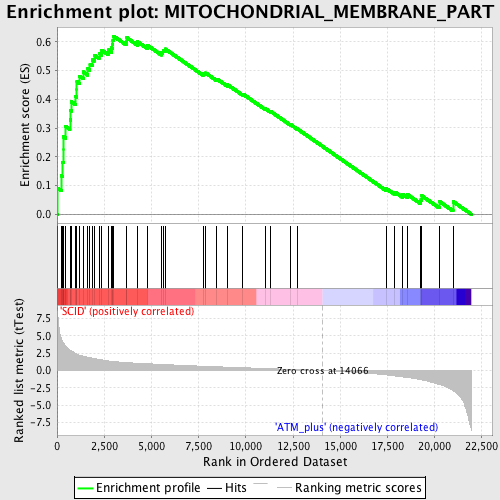
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | MITOCHONDRIAL_MEMBRANE_PART |
| Enrichment Score (ES) | 0.61963975 |
| Normalized Enrichment Score (NES) | 2.1091847 |
| Nominal p-value | 0.0 |
| FDR q-value | 6.2218955E-4 |
| FWER p-Value | 0.0060 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM50 | 1441819_x_at 1455456_a_at | 19 | 7.985 | 0.0906 | Yes | ||
| 2 | NDUFS7 | 1424313_a_at 1451312_at | 219 | 4.633 | 0.1346 | Yes | ||
| 3 | ATP5G1 | 1416020_a_at 1444874_at | 272 | 4.304 | 0.1815 | Yes | ||
| 4 | TIMM10 | 1417499_at | 323 | 3.974 | 0.2248 | Yes | ||
| 5 | ATP5G2 | 1415980_at 1456128_at | 325 | 3.967 | 0.2702 | Yes | ||
| 6 | ATP5E | 1416567_s_at | 446 | 3.593 | 0.3059 | Yes | ||
| 7 | NDUFA9 | 1416663_at | 683 | 2.972 | 0.3291 | Yes | ||
| 8 | NDUFV1 | 1415966_a_at 1415967_at 1456015_x_at | 723 | 2.891 | 0.3605 | Yes | ||
| 9 | BCS1L | 1451541_at | 759 | 2.821 | 0.3912 | Yes | ||
| 10 | UQCRC1 | 1428782_a_at | 988 | 2.464 | 0.4090 | Yes | ||
| 11 | NDUFB6 | 1434056_a_at 1434057_at | 1047 | 2.394 | 0.4338 | Yes | ||
| 12 | TIMM13 | 1455211_a_at | 1050 | 2.391 | 0.4611 | Yes | ||
| 13 | ATP5O | 1416278_a_at | 1186 | 2.221 | 0.4804 | Yes | ||
| 14 | NDUFS8 | 1423907_a_at 1423908_at 1434212_at 1434213_x_at 1434579_x_at 1455283_x_at | 1384 | 2.070 | 0.4951 | Yes | ||
| 15 | TIMM23 | 1416485_at | 1627 | 1.937 | 0.5062 | Yes | ||
| 16 | COX6B2 | 1435275_at | 1740 | 1.878 | 0.5226 | Yes | ||
| 17 | NDUFA6 | 1448427_at | 1879 | 1.795 | 0.5369 | Yes | ||
| 18 | TOMM22 | 1429103_at 1434420_x_at | 1967 | 1.747 | 0.5529 | Yes | ||
| 19 | NDUFS2 | 1451096_at | 2223 | 1.610 | 0.5597 | Yes | ||
| 20 | ATP5B | 1416829_at | 2361 | 1.539 | 0.5711 | Yes | ||
| 21 | NDUFA2 | 1417368_s_at | 2704 | 1.402 | 0.5715 | Yes | ||
| 22 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 2886 | 1.346 | 0.5787 | Yes | ||
| 23 | NDUFS3 | 1423737_at | 2935 | 1.330 | 0.5917 | Yes | ||
| 24 | NDUFA1 | 1422241_a_at | 2955 | 1.324 | 0.6060 | Yes | ||
| 25 | TIMM9 | 1428010_at 1449886_a_at | 2988 | 1.315 | 0.6196 | Yes | ||
| 26 | TIMM17A | 1426256_at | 3657 | 1.154 | 0.6023 | No | ||
| 27 | TIMM8B | 1431665_a_at | 3694 | 1.148 | 0.6139 | No | ||
| 28 | ATP5D | 1423716_s_at 1456580_s_at | 4261 | 1.052 | 0.6000 | No | ||
| 29 | ATP5C1 | 1416058_s_at 1438809_at | 4775 | 0.992 | 0.5880 | No | ||
| 30 | NDUFS1 | 1425143_a_at 1453354_at | 5524 | 0.892 | 0.5640 | No | ||
| 31 | MFN2 | 1441424_at 1448131_at 1451900_at | 5608 | 0.882 | 0.5703 | No | ||
| 32 | NDUFS4 | 1418117_at 1438166_x_at 1444464_at 1448959_at 1459760_at | 5725 | 0.865 | 0.5749 | No | ||
| 33 | ATP5G3 | 1454661_at | 7732 | 0.627 | 0.4905 | No | ||
| 34 | FIS1 | 1416764_at 1443819_x_at | 7866 | 0.614 | 0.4914 | No | ||
| 35 | TIMM17B | 1438656_x_at 1460685_at | 8465 | 0.553 | 0.4705 | No | ||
| 36 | NDUFA13 | 1430713_s_at | 9007 | 0.499 | 0.4515 | No | ||
| 37 | SURF1 | 1450561_a_at | 9843 | 0.417 | 0.4181 | No | ||
| 38 | BNIP3 | 1422470_at | 11032 | 0.306 | 0.3673 | No | ||
| 39 | OXA1L | 1423738_at 1451088_a_at 1456845_at | 11302 | 0.281 | 0.3582 | No | ||
| 40 | UQCRH | 1452133_at 1453229_s_at | 12385 | 0.175 | 0.3108 | No | ||
| 41 | SDHA | 1426688_at 1426689_s_at 1445317_at | 12709 | 0.145 | 0.2977 | No | ||
| 42 | COX15 | 1426693_x_at 1437982_x_at 1452146_a_at 1460376_a_at | 17448 | -0.622 | 0.0884 | No | ||
| 43 | PPOX | 1416618_at 1441862_at 1448065_at | 17894 | -0.769 | 0.0769 | No | ||
| 44 | UQCRB | 1416337_at 1455997_a_at | 18297 | -0.912 | 0.0689 | No | ||
| 45 | RHOT1 | 1423533_a_at 1440853_at | 18559 | -0.996 | 0.0684 | No | ||
| 46 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 19271 | -1.302 | 0.0509 | No | ||
| 47 | NNT | 1416105_at 1432608_at 1456573_x_at | 19316 | -1.330 | 0.0641 | No | ||
| 48 | OPA1 | 1418768_at 1434890_at 1449214_a_at | 20236 | -1.993 | 0.0449 | No | ||
| 49 | RHOT2 | 1426822_at | 20972 | -2.853 | 0.0440 | No |

