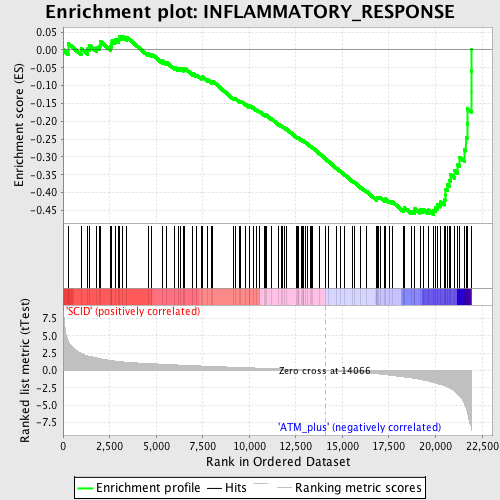
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | INFLAMMATORY_RESPONSE |
| Enrichment Score (ES) | -0.4610204 |
| Normalized Enrichment Score (NES) | -1.7189342 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2021344 |
| FWER p-Value | 0.971 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 1418901_at 1427844_a_at | 270 | 4.311 | 0.0182 | No | ||
| 2 | SCYE1 | 1416486_at | 965 | 2.494 | 0.0041 | No | ||
| 3 | TPST1 | 1421733_a_at | 1325 | 2.112 | 0.0026 | No | ||
| 4 | PTX3 | 1418666_at 1456903_at | 1423 | 2.040 | 0.0126 | No | ||
| 5 | IL5 | 1425546_a_at 1442049_at 1450550_at | 1813 | 1.839 | 0.0079 | No | ||
| 6 | AHSG | 1455093_a_at | 1979 | 1.741 | 0.0127 | No | ||
| 7 | ADORA2A | 1427519_at 1460710_at | 2026 | 1.709 | 0.0227 | No | ||
| 8 | TACR1 | 1422282_at | 2549 | 1.453 | 0.0091 | No | ||
| 9 | CCL3 | 1419561_at | 2591 | 1.439 | 0.0174 | No | ||
| 10 | AGER | 1420428_at 1441958_s_at | 2624 | 1.428 | 0.0260 | No | ||
| 11 | C3AR1 | 1419482_at 1419483_at 1442082_at | 2787 | 1.376 | 0.0284 | No | ||
| 12 | PTAFR | 1427871_at 1427872_at | 2975 | 1.319 | 0.0292 | No | ||
| 13 | CXCL1 | 1419209_at 1441855_x_at 1457644_s_at | 3009 | 1.309 | 0.0369 | No | ||
| 14 | PLA2G2E | 1420674_at | 3170 | 1.261 | 0.0386 | No | ||
| 15 | PLA2G7 | 1430700_a_at | 3424 | 1.207 | 0.0355 | No | ||
| 16 | HDAC9 | 1421306_a_at 1434572_at | 4559 | 1.014 | -0.0092 | No | ||
| 17 | IL9 | 1450565_at | 4771 | 0.992 | -0.0119 | No | ||
| 18 | C2 | 1416051_at 1441912_x_at 1457664_x_at | 5321 | 0.921 | -0.0305 | No | ||
| 19 | IL18RAP | 1421291_at 1456545_at | 5542 | 0.890 | -0.0343 | No | ||
| 20 | IL1RAP | 1421843_at 1439697_at 1442614_at 1449585_at | 5971 | 0.835 | -0.0480 | No | ||
| 21 | CCR5 | 1422259_a_at 1422260_x_at 1424727_at | 6179 | 0.807 | -0.0517 | No | ||
| 22 | SCG2 | 1450708_at | 6330 | 0.789 | -0.0530 | No | ||
| 23 | GHSR | 1446756_at | 6469 | 0.771 | -0.0538 | No | ||
| 24 | CXCL2 | 1449984_at | 6533 | 0.764 | -0.0513 | No | ||
| 25 | GHRL | 1448980_at | 6974 | 0.714 | -0.0664 | No | ||
| 26 | ABCF1 | 1420157_s_at 1420158_s_at 1426921_at 1427444_at 1452236_at 1452429_s_at | 7177 | 0.691 | -0.0708 | No | ||
| 27 | CCL24 | 1450488_at | 7417 | 0.661 | -0.0770 | No | ||
| 28 | ELF3 | 1416916_at | 7458 | 0.656 | -0.0742 | No | ||
| 29 | LBP | 1448550_at | 7751 | 0.625 | -0.0832 | No | ||
| 30 | KNG1 | 1416676_at 1426045_at | 7968 | 0.603 | -0.0888 | No | ||
| 31 | APCS | 1419059_at | 8037 | 0.596 | -0.0877 | No | ||
| 32 | RIPK2 | 1421236_at 1450173_at | 9165 | 0.482 | -0.1359 | No | ||
| 33 | PLA2G2D | 1418987_at 1454157_a_at | 9262 | 0.475 | -0.1369 | No | ||
| 34 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 9497 | 0.451 | -0.1444 | No | ||
| 35 | IFNA2 | 1423028_at | 9546 | 0.446 | -0.1435 | No | ||
| 36 | CCL11 | 1417789_at | 9810 | 0.420 | -0.1525 | No | ||
| 37 | AOX1 | 1419435_at | 10031 | 0.398 | -0.1598 | No | ||
| 38 | IL13 | 1420802_at | 10032 | 0.398 | -0.1570 | No | ||
| 39 | FPRL1 | 1421661_at 1450583_s_at | 10203 | 0.383 | -0.1620 | No | ||
| 40 | ORM2 | 1420438_at | 10394 | 0.366 | -0.1682 | No | ||
| 41 | IL20 | 1421608_at | 10525 | 0.354 | -0.1716 | No | ||
| 42 | NFATC4 | 1423380_s_at 1432821_at 1454369_a_at | 10798 | 0.325 | -0.1818 | No | ||
| 43 | ORM1 | 1451054_at | 10884 | 0.317 | -0.1834 | No | ||
| 44 | CHRNA7 | 1440681_at 1450299_at | 10906 | 0.316 | -0.1821 | No | ||
| 45 | ADORA1 | 1427331_at 1435495_at | 11175 | 0.294 | -0.1923 | No | ||
| 46 | CFHR1 | 1419436_at | 11577 | 0.251 | -0.2089 | No | ||
| 47 | IL8RB | 1421734_at | 11705 | 0.238 | -0.2130 | No | ||
| 48 | BLNK | 1451780_at | 11797 | 0.229 | -0.2156 | No | ||
| 49 | F11R | 1424595_at 1436374_x_at | 11896 | 0.218 | -0.2185 | No | ||
| 50 | CCR2 | 1421186_at 1421187_at 1421188_at 1460067_at | 11984 | 0.210 | -0.2210 | No | ||
| 51 | SELE | 1421712_at | 12540 | 0.161 | -0.2453 | No | ||
| 52 | CRP | 1421946_at | 12604 | 0.155 | -0.2471 | No | ||
| 53 | EDG3 | 1437173_at 1438658_a_at 1460661_at | 12616 | 0.154 | -0.2465 | No | ||
| 54 | AOAH | 1450764_at 1460156_at | 12651 | 0.150 | -0.2470 | No | ||
| 55 | CCL4 | 1421578_at | 12786 | 0.139 | -0.2522 | No | ||
| 56 | TNFAIP6 | 1418424_at | 12877 | 0.128 | -0.2554 | No | ||
| 57 | ALOX5AP | 1452016_at 1459234_at | 12889 | 0.128 | -0.2550 | No | ||
| 58 | IL1A | 1421473_at | 12916 | 0.125 | -0.2553 | No | ||
| 59 | AOC3 | 1449396_at | 12999 | 0.116 | -0.2582 | No | ||
| 60 | MEFV | 1460283_at | 13113 | 0.104 | -0.2627 | No | ||
| 61 | NOX4 | 1419161_a_at 1451827_a_at | 13309 | 0.080 | -0.2710 | No | ||
| 62 | CCR1 | 1419609_at 1419610_at | 13355 | 0.076 | -0.2725 | No | ||
| 63 | CCL20 | 1422029_at | 13409 | 0.070 | -0.2745 | No | ||
| 64 | CXCL11 | 1419697_at 1419698_at | 13752 | 0.033 | -0.2899 | No | ||
| 65 | CCL5 | 1418126_at | 14078 | -0.001 | -0.3048 | No | ||
| 66 | MBL2 | 1418787_at | 14249 | -0.022 | -0.3124 | No | ||
| 67 | CHST2 | 1422758_at 1460070_at | 14688 | -0.077 | -0.3319 | No | ||
| 68 | XCR1 | 1422294_at | 14891 | -0.102 | -0.3405 | No | ||
| 69 | IL10RB | 1419455_at | 15132 | -0.134 | -0.3505 | No | ||
| 70 | HRH1 | 1419210_at 1438494_at | 15548 | -0.196 | -0.3681 | No | ||
| 71 | LTB4R | 1420407_at | 15637 | -0.209 | -0.3707 | No | ||
| 72 | KLRG1 | 1420788_at | 15996 | -0.271 | -0.3852 | No | ||
| 73 | MGLL | 1426785_s_at 1431331_at 1450391_a_at 1453836_a_at | 16281 | -0.325 | -0.3959 | No | ||
| 74 | NFKB1 | 1427705_a_at 1442949_at | 16839 | -0.453 | -0.4182 | No | ||
| 75 | CD40 | 1439221_s_at 1449473_s_at 1460415_a_at | 16848 | -0.457 | -0.4153 | No | ||
| 76 | SIGIRR | 1449163_at | 16868 | -0.462 | -0.4129 | No | ||
| 77 | CDO1 | 1448842_at | 16927 | -0.476 | -0.4122 | No | ||
| 78 | CCR3 | 1422957_at | 17019 | -0.499 | -0.4128 | No | ||
| 79 | CD40LG | 1422283_at | 17262 | -0.570 | -0.4198 | No | ||
| 80 | CXCL10 | 1418930_at | 17296 | -0.580 | -0.4172 | No | ||
| 81 | CXCL9 | 1418652_at 1456907_at | 17549 | -0.659 | -0.4241 | No | ||
| 82 | AIF1 | 1418204_s_at | 17668 | -0.693 | -0.4246 | No | ||
| 83 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 18298 | -0.913 | -0.4470 | No | ||
| 84 | PRDX5 | 1416381_a_at | 18318 | -0.918 | -0.4413 | No | ||
| 85 | CYBB | 1422978_at 1436778_at 1436779_at | 18699 | -1.038 | -0.4514 | No | ||
| 86 | CCR4 | 1421655_a_at | 18874 | -1.109 | -0.4515 | Yes | ||
| 87 | IRAK2 | 1436507_at | 18879 | -1.112 | -0.4438 | Yes | ||
| 88 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 19167 | -1.249 | -0.4481 | Yes | ||
| 89 | GPR68 | 1455000_at | 19331 | -1.339 | -0.4460 | Yes | ||
| 90 | ANXA1 | 1448213_at | 19616 | -1.511 | -0.4483 | Yes | ||
| 91 | NFE2L1 | 1416331_a_at | 19894 | -1.715 | -0.4489 | Yes | ||
| 92 | CXCR4 | 1448710_at | 19996 | -1.803 | -0.4407 | Yes | ||
| 93 | HDAC4 | 1436758_at 1447566_at 1454693_at | 20114 | -1.896 | -0.4326 | Yes | ||
| 94 | RELA | 1419536_a_at | 20278 | -2.013 | -0.4258 | Yes | ||
| 95 | LY75 | 1449328_at | 20468 | -2.172 | -0.4191 | Yes | ||
| 96 | ALOX15 | 1420338_at | 20542 | -2.258 | -0.4064 | Yes | ||
| 97 | CD97 | 1418394_a_at | 20547 | -2.260 | -0.3906 | Yes | ||
| 98 | NMI | 1425719_a_at | 20625 | -2.363 | -0.3773 | Yes | ||
| 99 | CCL22 | 1417925_at | 20749 | -2.506 | -0.3652 | Yes | ||
| 100 | NFRKB | 1434753_at 1445551_at | 20817 | -2.610 | -0.3498 | Yes | ||
| 101 | PARP4 | 1441026_at | 21041 | -2.979 | -0.3388 | Yes | ||
| 102 | TNFRSF1A | 1417291_at | 21202 | -3.374 | -0.3222 | Yes | ||
| 103 | FOS | 1423100_at | 21307 | -3.658 | -0.3011 | Yes | ||
| 104 | HDAC5 | 1415743_at | 21541 | -4.626 | -0.2789 | Yes | ||
| 105 | HDAC7A | 1420812_at 1420813_at | 21637 | -5.344 | -0.2454 | Yes | ||
| 106 | NFATC3 | 1419976_s_at 1452497_a_at | 21694 | -5.798 | -0.2069 | Yes | ||
| 107 | TGFB1 | 1420653_at 1445360_at | 21716 | -6.029 | -0.1651 | Yes | ||
| 108 | S100A9 | 1448756_at | 21916 | -8.114 | -0.1166 | Yes | ||
| 109 | CCR7 | 1423466_at | 21917 | -8.151 | -0.0588 | Yes | ||
| 110 | S100A8 | 1419394_s_at | 21930 | -8.411 | 0.0003 | Yes |

