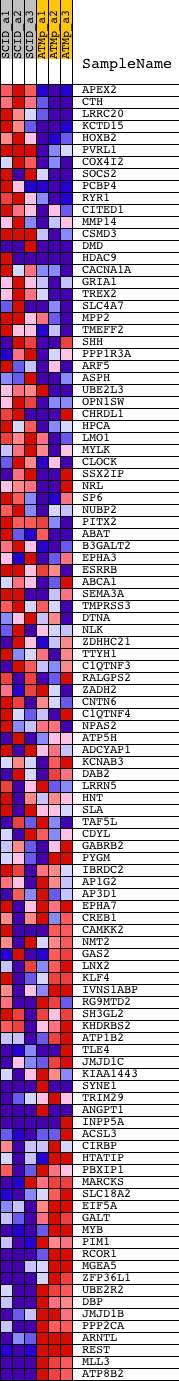
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
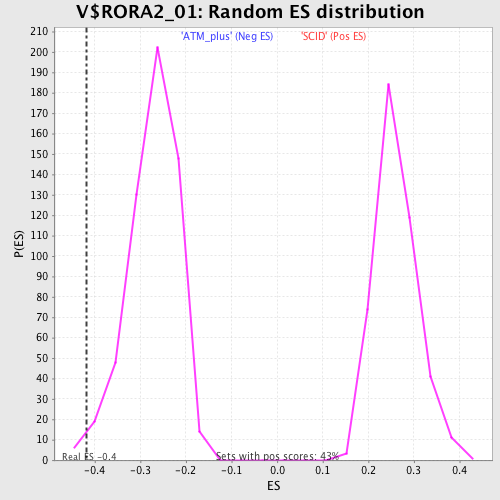
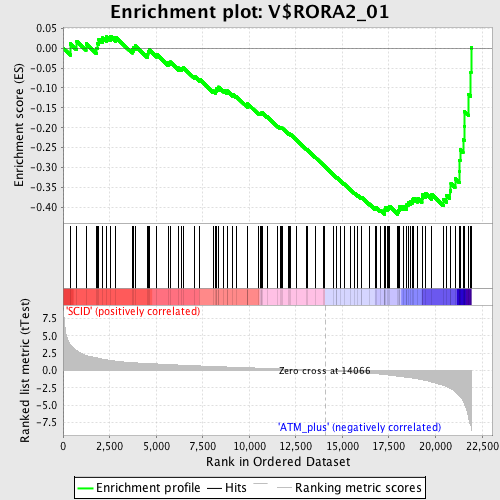
| GeneSet | V$RORA2_01 |
| Enrichment Score (ES) | -0.41802633 |
| Normalized Enrichment Score (NES) | -1.5384014 |
| Nominal p-value | 0.01058201 |
| FDR q-value | 0.120168746 |
| FWER p-Value | 0.849 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | APEX2 | 1424322_at 1425954_a_at | 384 | 3.763 | 0.0116 | No | ||
| 2 | CTH | 1426243_at | 737 | 2.869 | 0.0176 | No | ||
| 3 | LRRC20 | 1438422_at | 1231 | 2.187 | 0.0120 | No | ||
| 4 | KCTD15 | 1435339_at | 1802 | 1.845 | 0.0002 | No | ||
| 5 | HOXB2 | 1444204_at 1449397_at | 1854 | 1.808 | 0.0118 | No | ||
| 6 | PVRL1 | 1438111_at 1438421_at 1440131_at 1446488_at 1450819_at | 1919 | 1.772 | 0.0226 | No | ||
| 7 | COX4I2 | 1421373_at | 2130 | 1.654 | 0.0258 | No | ||
| 8 | SOCS2 | 1418507_s_at 1438470_at 1441476_at 1442586_at 1446085_at 1449109_at | 2336 | 1.549 | 0.0284 | No | ||
| 9 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 2544 | 1.455 | 0.0302 | No | ||
| 10 | RYR1 | 1427306_at 1457347_at | 2826 | 1.360 | 0.0279 | No | ||
| 11 | CITED1 | 1449031_at | 3704 | 1.147 | -0.0034 | No | ||
| 12 | MMP14 | 1416572_at 1448383_at | 3795 | 1.132 | 0.0012 | No | ||
| 13 | CSMD3 | 1438272_at 1438572_at 1454245_at | 3884 | 1.115 | 0.0058 | No | ||
| 14 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 4532 | 1.018 | -0.0159 | No | ||
| 15 | HDAC9 | 1421306_a_at 1434572_at | 4559 | 1.014 | -0.0093 | No | ||
| 16 | CACNA1A | 1430408_at 1444455_at 1450510_a_at 1458312_at 1459996_at | 4618 | 1.009 | -0.0041 | No | ||
| 17 | GRIA1 | 1435239_at 1448972_at 1458285_at | 5021 | 0.963 | -0.0151 | No | ||
| 18 | TREX2 | 1449367_at | 5642 | 0.877 | -0.0367 | No | ||
| 19 | SLC4A7 | 1455876_at 1457528_at | 5743 | 0.862 | -0.0346 | No | ||
| 20 | MPP2 | 1449173_at | 6209 | 0.803 | -0.0497 | No | ||
| 21 | TMEFF2 | 1419073_at 1441598_at | 6349 | 0.787 | -0.0500 | No | ||
| 22 | SHH | 1427571_at 1436869_at | 6447 | 0.774 | -0.0484 | No | ||
| 23 | PPP1R3A | 1422108_at | 7047 | 0.705 | -0.0704 | No | ||
| 24 | ARF5 | 1422807_at | 7328 | 0.671 | -0.0780 | No | ||
| 25 | ASPH | 1420959_at 1425274_at 1425275_at 1426015_s_at 1436455_at 1450058_at | 8073 | 0.591 | -0.1075 | No | ||
| 26 | UBE2L3 | 1417907_at 1417908_s_at 1448879_at 1448880_at | 8198 | 0.580 | -0.1087 | No | ||
| 27 | OPN1SW | 1418552_at 1449132_at | 8230 | 0.577 | -0.1057 | No | ||
| 28 | CHRDL1 | 1421295_at 1424945_at 1434201_at 1456722_at | 8233 | 0.577 | -0.1013 | No | ||
| 29 | HPCA | 1425833_a_at 1450930_at | 8343 | 0.566 | -0.1019 | No | ||
| 30 | LMO1 | 1418478_at | 8347 | 0.566 | -0.0977 | No | ||
| 31 | MYLK | 1425504_at 1425505_at 1425506_at | 8626 | 0.537 | -0.1062 | No | ||
| 32 | CLOCK | 1418659_at 1435775_at 1439314_at | 8817 | 0.519 | -0.1109 | No | ||
| 33 | SSX2IP | 1417514_at 1420185_at 1448743_at 1449764_x_at | 8829 | 0.518 | -0.1074 | No | ||
| 34 | NRL | 1450946_at | 9121 | 0.487 | -0.1170 | No | ||
| 35 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 9286 | 0.473 | -0.1208 | No | ||
| 36 | NUBP2 | 1416512_at 1459842_x_at | 9902 | 0.412 | -0.1458 | No | ||
| 37 | PITX2 | 1424797_a_at 1450482_a_at | 9912 | 0.411 | -0.1430 | No | ||
| 38 | ABAT | 1433855_at | 9914 | 0.411 | -0.1399 | No | ||
| 39 | B3GALT2 | 1423084_at 1437433_at 1437644_at | 10481 | 0.357 | -0.1631 | No | ||
| 40 | EPHA3 | 1425574_at 1425575_at 1426057_a_at 1443273_at | 10589 | 0.346 | -0.1653 | No | ||
| 41 | ESRRB | 1422986_at 1436926_at 1441921_x_at | 10646 | 0.341 | -0.1652 | No | ||
| 42 | ABCA1 | 1421839_at 1421840_at 1450392_at | 10652 | 0.341 | -0.1628 | No | ||
| 43 | SEMA3A | 1420416_at 1420417_at 1449865_at | 10699 | 0.336 | -0.1623 | No | ||
| 44 | TMPRSS3 | 1421714_at | 10955 | 0.312 | -0.1716 | No | ||
| 45 | DTNA | 1419223_a_at 1425292_at 1426066_a_at 1427588_a_at 1429768_at 1447280_at 1456069_at 1458908_at | 11539 | 0.256 | -0.1963 | No | ||
| 46 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 11670 | 0.242 | -0.2004 | No | ||
| 47 | ZDHHC21 | 1429488_at | 11717 | 0.238 | -0.2007 | No | ||
| 48 | TTYH1 | 1422694_at 1422696_at 1426617_a_at | 11757 | 0.232 | -0.2006 | No | ||
| 49 | C1QTNF3 | 1422606_at | 12113 | 0.199 | -0.2154 | No | ||
| 50 | RALGPS2 | 1417230_at 1428789_at 1431704_a_at | 12143 | 0.196 | -0.2152 | No | ||
| 51 | ZADH2 | 1451277_at | 12229 | 0.189 | -0.2176 | No | ||
| 52 | CNTN6 | 1422649_at | 12549 | 0.159 | -0.2310 | No | ||
| 53 | C1QTNF4 | 1417050_at 1441648_at | 13084 | 0.107 | -0.2546 | No | ||
| 54 | NPAS2 | 1421036_at 1421037_at | 13130 | 0.102 | -0.2559 | No | ||
| 55 | ATP5H | 1423676_at 1435112_a_at | 13568 | 0.052 | -0.2755 | No | ||
| 56 | ADCYAP1 | 1423427_at 1441778_at | 13995 | 0.008 | -0.2950 | No | ||
| 57 | KCNAB3 | 1421790_a_at 1423950_at | 14016 | 0.005 | -0.2958 | No | ||
| 58 | DAB2 | 1420498_a_at 1423805_at 1429693_at 1430604_a_at | 14520 | -0.056 | -0.3185 | No | ||
| 59 | LRRN5 | 1433469_at | 14698 | -0.079 | -0.3260 | No | ||
| 60 | HNT | 1426282_at 1426283_at 1458492_x_at | 14702 | -0.079 | -0.3255 | No | ||
| 61 | SLA | 1420818_at 1420819_at 1441761_at 1447813_x_at | 14873 | -0.100 | -0.3325 | No | ||
| 62 | TAF5L | 1448195_at | 15087 | -0.129 | -0.3413 | No | ||
| 63 | CDYL | 1418070_at 1418071_s_at 1448935_at | 15448 | -0.183 | -0.3563 | No | ||
| 64 | GABRB2 | 1450319_at | 15670 | -0.214 | -0.3648 | No | ||
| 65 | PYGM | 1448602_at | 15834 | -0.240 | -0.3704 | No | ||
| 66 | IBRDC2 | 1425282_at 1439153_at 1443252_at 1456834_at | 16015 | -0.274 | -0.3765 | No | ||
| 67 | AP1G2 | 1419113_at | 16047 | -0.281 | -0.3758 | No | ||
| 68 | AP3D1 | 1417964_at 1445280_at | 16453 | -0.359 | -0.3915 | No | ||
| 69 | EPHA7 | 1427528_a_at 1443485_at 1445635_at 1451991_at 1452380_at | 16772 | -0.438 | -0.4027 | No | ||
| 70 | CREB1 | 1421582_a_at 1421583_at 1423402_at 1428755_at 1452529_a_at 1452901_at | 16806 | -0.445 | -0.4008 | No | ||
| 71 | CAMKK2 | 1424474_a_at 1424475_at 1424476_at 1455401_at | 17025 | -0.500 | -0.4069 | No | ||
| 72 | NMT2 | 1423581_at 1440433_at 1444387_at 1457107_at | 17269 | -0.572 | -0.4136 | Yes | ||
| 73 | GAS2 | 1429147_at 1435834_at 1438564_at 1450112_a_at | 17276 | -0.573 | -0.4094 | Yes | ||
| 74 | LNX2 | 1460249_at | 17284 | -0.575 | -0.4053 | Yes | ||
| 75 | KLF4 | 1417394_at 1417395_at | 17293 | -0.578 | -0.4012 | Yes | ||
| 76 | IVNS1ABP | 1420961_a_at 1425718_a_at 1450084_s_at | 17445 | -0.621 | -0.4033 | Yes | ||
| 77 | RG9MTD2 | 1427375_at 1435035_at 1447781_s_at | 17478 | -0.632 | -0.3999 | Yes | ||
| 78 | SH3GL2 | 1418791_at 1418792_at 1449228_at | 17528 | -0.654 | -0.3971 | Yes | ||
| 79 | KHDRBS2 | 1422117_s_at 1445110_at | 17976 | -0.795 | -0.4114 | Yes | ||
| 80 | ATP1B2 | 1422009_at 1435148_at | 18018 | -0.812 | -0.4070 | Yes | ||
| 81 | TLE4 | 1425650_at 1430384_at 1450853_at | 18056 | -0.832 | -0.4022 | Yes | ||
| 82 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 18083 | -0.838 | -0.3969 | Yes | ||
| 83 | KIAA1443 | 1434636_at | 18256 | -0.899 | -0.3979 | Yes | ||
| 84 | SYNE1 | 1421545_a_at 1455225_at 1455493_at | 18443 | -0.963 | -0.3989 | Yes | ||
| 85 | TRIM29 | 1424162_at | 18463 | -0.968 | -0.3923 | Yes | ||
| 86 | ANGPT1 | 1421441_at 1438936_s_at 1438937_x_at 1439066_at 1446656_at 1450717_at 1457381_at | 18530 | -0.991 | -0.3876 | Yes | ||
| 87 | INPP5A | 1433605_at 1446114_at | 18646 | -1.023 | -0.3850 | Yes | ||
| 88 | ACSL3 | 1428386_at 1428387_at 1452771_s_at | 18745 | -1.055 | -0.3813 | Yes | ||
| 89 | CIRBP | 1416332_at | 18829 | -1.090 | -0.3767 | Yes | ||
| 90 | HTATIP | 1433980_at 1433981_s_at | 19053 | -1.191 | -0.3777 | Yes | ||
| 91 | PBXIP1 | 1451132_at | 19275 | -1.304 | -0.3777 | Yes | ||
| 92 | MARCKS | 1415971_at 1415972_at 1430311_at 1437034_x_at 1456028_x_at | 19290 | -1.314 | -0.3682 | Yes | ||
| 93 | SLC18A2 | 1437079_at | 19475 | -1.425 | -0.3656 | Yes | ||
| 94 | EIF5A | 1437859_x_at 1451470_s_at | 19803 | -1.648 | -0.3678 | Yes | ||
| 95 | GALT | 1450187_a_at | 20432 | -2.136 | -0.3800 | Yes | ||
| 96 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 20588 | -2.317 | -0.3692 | Yes | ||
| 97 | PIM1 | 1423006_at 1435458_at | 20778 | -2.545 | -0.3581 | Yes | ||
| 98 | RCOR1 | 1434336_s_at 1437545_at 1454932_at | 20828 | -2.619 | -0.3401 | Yes | ||
| 99 | MGEA5 | 1422900_at 1422901_at 1422902_s_at 1433654_at 1439081_at | 21049 | -2.990 | -0.3270 | Yes | ||
| 100 | ZFP36L1 | 1422528_a_at 1450644_at | 21283 | -3.593 | -0.3099 | Yes | ||
| 101 | UBE2R2 | 1415768_a_at 1443742_x_at 1444460_at | 21292 | -3.617 | -0.2822 | Yes | ||
| 102 | DBP | 1418174_at 1438211_s_at | 21322 | -3.702 | -0.2549 | Yes | ||
| 103 | JMJD1B | 1428320_at 1438508_at 1458417_at | 21480 | -4.281 | -0.2289 | Yes | ||
| 104 | PPP2CA | 1417367_at 1444875_at 1456390_at | 21548 | -4.655 | -0.1959 | Yes | ||
| 105 | ARNTL | 1425099_a_at 1446544_at 1457740_at | 21561 | -4.763 | -0.1596 | Yes | ||
| 106 | REST | 1425564_at 1425565_at 1428227_at | 21784 | -6.888 | -0.1164 | Yes | ||
| 107 | MLL3 | 1427150_at 1434178_at 1434179_at 1457193_at | 21897 | -7.909 | -0.0603 | Yes | ||
| 108 | ATP8B2 | 1434026_at 1441699_at 1452450_at | 21910 | -8.016 | 0.0012 | Yes |