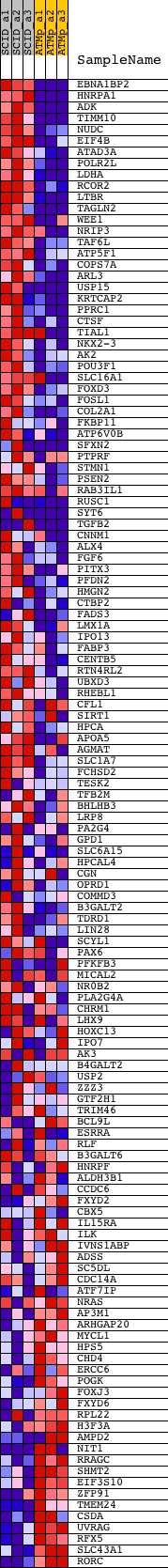
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
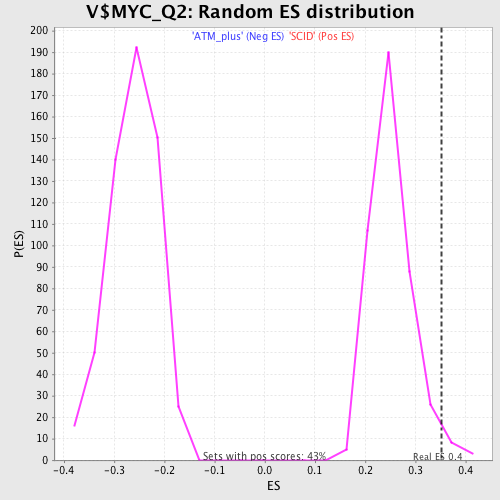
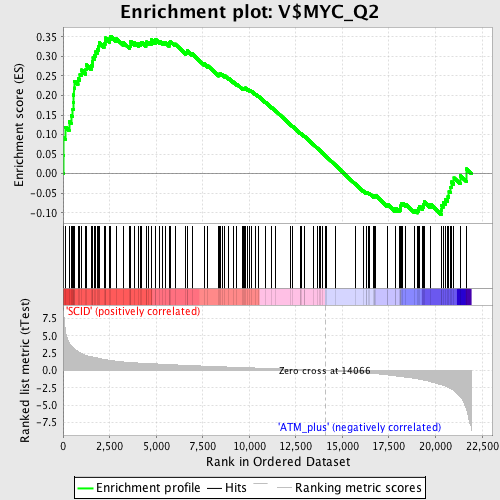
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.35199848 |
| Normalized Enrichment Score (NES) | 1.398347 |
| Nominal p-value | 0.023419203 |
| FDR q-value | 0.14404917 |
| FWER p-Value | 0.991 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EBNA1BP2 | 1428315_at 1437512_x_at 1439898_at 1457668_x_at | 14 | 8.047 | 0.0462 | Yes | ||
| 2 | HNRPA1 | 1423531_a_at 1430020_x_at 1436549_a_at 1442758_at 1446038_at 1455305_x_at 1455740_at | 26 | 7.887 | 0.0917 | Yes | ||
| 3 | ADK | 1416319_at 1421767_at 1438292_x_at 1442615_at 1445402_at 1446068_at 1446675_at 1456960_at 1459645_at | 135 | 5.501 | 0.1188 | Yes | ||
| 4 | TIMM10 | 1417499_at | 323 | 3.974 | 0.1333 | Yes | ||
| 5 | NUDC | 1419471_a_at | 433 | 3.613 | 0.1494 | Yes | ||
| 6 | EIF4B | 1426378_at 1426379_at 1426380_at | 523 | 3.366 | 0.1649 | Yes | ||
| 7 | ATAD3A | 1422461_at 1437343_x_at 1438178_x_at 1456541_x_at | 547 | 3.304 | 0.1831 | Yes | ||
| 8 | POLR2L | 1428296_at | 562 | 3.269 | 0.2015 | Yes | ||
| 9 | LDHA | 1419737_a_at | 584 | 3.215 | 0.2193 | Yes | ||
| 10 | RCOR2 | 1417302_at | 627 | 3.122 | 0.2356 | Yes | ||
| 11 | LTBR | 1416435_at | 801 | 2.755 | 0.2437 | Yes | ||
| 12 | TAGLN2 | 1426529_a_at | 904 | 2.585 | 0.2541 | Yes | ||
| 13 | WEE1 | 1416773_at 1416774_at | 973 | 2.481 | 0.2654 | Yes | ||
| 14 | NRIP3 | 1418104_at 1431597_a_at 1448954_at | 1223 | 2.192 | 0.2667 | Yes | ||
| 15 | TAF6L | 1452600_at | 1242 | 2.173 | 0.2786 | Yes | ||
| 16 | ATP5F1 | 1426742_at 1433562_s_at | 1524 | 1.992 | 0.2773 | Yes | ||
| 17 | COPS7A | 1423245_at 1429078_a_at | 1568 | 1.971 | 0.2868 | Yes | ||
| 18 | ARL3 | 1450706_a_at | 1603 | 1.950 | 0.2966 | Yes | ||
| 19 | USP15 | 1436891_at 1446186_at 1454036_a_at 1455842_x_at | 1693 | 1.905 | 0.3036 | Yes | ||
| 20 | KRTCAP2 | 1417058_a_at 1417059_at | 1758 | 1.870 | 0.3116 | Yes | ||
| 21 | PPRC1 | 1426381_at | 1829 | 1.824 | 0.3190 | Yes | ||
| 22 | CTSF | 1449794_x_at 1451019_at | 1914 | 1.776 | 0.3255 | Yes | ||
| 23 | TIAL1 | 1421148_a_at 1426352_s_at 1446713_at 1452821_at 1455675_a_at 1455676_x_at | 1928 | 1.769 | 0.3352 | Yes | ||
| 24 | NKX2-3 | 1425827_at 1427741_x_at 1452528_a_at | 2199 | 1.619 | 0.3323 | Yes | ||
| 25 | AK2 | 1448450_at 1448451_at | 2254 | 1.592 | 0.3390 | Yes | ||
| 26 | POU3F1 | 1422068_at 1460038_at | 2274 | 1.582 | 0.3474 | Yes | ||
| 27 | SLC16A1 | 1415802_at 1446274_at | 2511 | 1.470 | 0.3451 | Yes | ||
| 28 | FOXD3 | 1422210_at | 2547 | 1.454 | 0.3520 | Yes | ||
| 29 | FOSL1 | 1417487_at 1417488_at | 2872 | 1.348 | 0.3450 | No | ||
| 30 | COL2A1 | 1450567_a_at | 3244 | 1.244 | 0.3352 | No | ||
| 31 | FKBP11 | 1417267_s_at 1425839_at 1425938_a_at | 3586 | 1.169 | 0.3264 | No | ||
| 32 | ATP6V0B | 1416769_s_at 1425448_x_at 1437013_x_at | 3611 | 1.163 | 0.3321 | No | ||
| 33 | SFXN2 | 1439966_x_at 1440786_x_at | 3614 | 1.163 | 0.3388 | No | ||
| 34 | PTPRF | 1420841_at 1420842_at 1420843_at | 3835 | 1.124 | 0.3352 | No | ||
| 35 | STMN1 | 1415849_s_at | 4037 | 1.088 | 0.3323 | No | ||
| 36 | PSEN2 | 1425869_a_at 1431542_at | 4141 | 1.072 | 0.3339 | No | ||
| 37 | RAB3IL1 | 1419991_at 1419992_x_at 1419993_at 1425133_s_at 1428391_at 1456442_at | 4220 | 1.059 | 0.3364 | No | ||
| 38 | RUSC1 | 1431137_at 1434743_x_at 1436014_a_at 1437991_x_at 1438017_at | 4452 | 1.026 | 0.3318 | No | ||
| 39 | SYT6 | 1420188_at 1426106_a_at 1449766_at 1449767_x_at | 4467 | 1.024 | 0.3371 | No | ||
| 40 | TGFB2 | 1423250_a_at 1438303_at 1446141_at 1450922_a_at 1450923_at | 4609 | 1.009 | 0.3366 | No | ||
| 41 | CNNM1 | 1420354_at 1441312_at 1445979_at | 4728 | 0.998 | 0.3370 | No | ||
| 42 | ALX4 | 1421737_at | 4731 | 0.997 | 0.3427 | No | ||
| 43 | FGF6 | 1427582_at | 4949 | 0.973 | 0.3384 | No | ||
| 44 | PITX3 | 1449917_at | 4951 | 0.972 | 0.3440 | No | ||
| 45 | PFDN2 | 1421950_at | 5200 | 0.937 | 0.3381 | No | ||
| 46 | HMGN2 | 1433507_a_at | 5345 | 0.917 | 0.3368 | No | ||
| 47 | CTBP2 | 1422887_a_at 1439823_at | 5476 | 0.898 | 0.3361 | No | ||
| 48 | FADS3 | 1418773_at 1435910_at 1449219_at | 5689 | 0.872 | 0.3315 | No | ||
| 49 | LMX1A | 1421554_at | 5713 | 0.867 | 0.3355 | No | ||
| 50 | IPO13 | 1433464_at | 5744 | 0.862 | 0.3391 | No | ||
| 51 | FABP3 | 1416023_at 1444844_at | 6009 | 0.831 | 0.3318 | No | ||
| 52 | CENTB5 | 1439021_at | 6584 | 0.758 | 0.3099 | No | ||
| 53 | RTN4RL2 | 1439573_at | 6661 | 0.750 | 0.3108 | No | ||
| 54 | UBXD3 | 1426007_a_at 1451906_at 1456385_x_at 1457581_at | 6671 | 0.748 | 0.3148 | No | ||
| 55 | RHEBL1 | 1447818_x_at 1451516_at | 6930 | 0.718 | 0.3071 | No | ||
| 56 | CFL1 | 1448346_at 1455138_x_at | 7577 | 0.644 | 0.2812 | No | ||
| 57 | SIRT1 | 1418640_at 1458538_at | 7772 | 0.623 | 0.2760 | No | ||
| 58 | HPCA | 1425833_a_at 1450930_at | 8343 | 0.566 | 0.2531 | No | ||
| 59 | APOA5 | 1417610_at | 8371 | 0.563 | 0.2552 | No | ||
| 60 | AGMAT | 1427214_at | 8426 | 0.557 | 0.2559 | No | ||
| 61 | SLC1A7 | 1456971_at | 8534 | 0.546 | 0.2542 | No | ||
| 62 | FCHSD2 | 1434260_at 1456949_at | 8690 | 0.531 | 0.2502 | No | ||
| 63 | TESK2 | 1426546_at | 8893 | 0.512 | 0.2439 | No | ||
| 64 | TFB2M | 1423441_at | 9155 | 0.484 | 0.2348 | No | ||
| 65 | BHLHB3 | 1421099_at | 9337 | 0.467 | 0.2292 | No | ||
| 66 | LRP8 | 1421459_a_at 1440882_at 1442347_at | 9641 | 0.435 | 0.2178 | No | ||
| 67 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 9676 | 0.432 | 0.2188 | No | ||
| 68 | GPD1 | 1416204_at 1439396_x_at 1448249_at 1456732_at | 9751 | 0.426 | 0.2179 | No | ||
| 69 | SLC6A15 | 1426712_at | 9773 | 0.424 | 0.2194 | No | ||
| 70 | HPCAL4 | 1433987_at | 9904 | 0.412 | 0.2158 | No | ||
| 71 | CGN | 1430329_at 1435155_at | 10006 | 0.400 | 0.2135 | No | ||
| 72 | OPRD1 | 1422121_at | 10103 | 0.392 | 0.2114 | No | ||
| 73 | COMMD3 | 1433539_at 1454642_a_at | 10321 | 0.372 | 0.2036 | No | ||
| 74 | B3GALT2 | 1423084_at 1437433_at 1437644_at | 10481 | 0.357 | 0.1984 | No | ||
| 75 | TDRD1 | 1421513_at | 10863 | 0.319 | 0.1828 | No | ||
| 76 | LIN28 | 1437752_at | 11191 | 0.292 | 0.1695 | No | ||
| 77 | SCYL1 | 1430354_x_at 1436804_s_at 1451051_a_at | 11405 | 0.270 | 0.1613 | No | ||
| 78 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 12224 | 0.189 | 0.1249 | No | ||
| 79 | PFKFB3 | 1416432_at 1419882_at 1456676_a_at | 12330 | 0.180 | 0.1211 | No | ||
| 80 | MICAL2 | 1438726_at 1455685_at | 12739 | 0.143 | 0.1032 | No | ||
| 81 | NR0B2 | 1449854_at | 12761 | 0.141 | 0.1031 | No | ||
| 82 | PLA2G4A | 1448558_a_at | 12813 | 0.137 | 0.1015 | No | ||
| 83 | CHRM1 | 1439611_at 1450833_at | 12965 | 0.120 | 0.0953 | No | ||
| 84 | LHX9 | 1419324_at 1431598_a_at 1441313_x_at | 12981 | 0.118 | 0.0953 | No | ||
| 85 | HOXC13 | 1425874_at | 13449 | 0.065 | 0.0743 | No | ||
| 86 | IPO7 | 1442511_at 1454955_at 1458165_at | 13657 | 0.044 | 0.0650 | No | ||
| 87 | AK3 | 1423717_at 1423718_at 1432436_a_at 1458812_at | 13766 | 0.031 | 0.0603 | No | ||
| 88 | B4GALT2 | 1418080_at 1448941_at | 13799 | 0.028 | 0.0590 | No | ||
| 89 | USP2 | 1417168_a_at 1417169_at | 13938 | 0.014 | 0.0527 | No | ||
| 90 | ZZZ3 | 1434332_at 1438487_s_at 1441748_at | 14101 | -0.004 | 0.0453 | No | ||
| 91 | GTF2H1 | 1453169_a_at | 14164 | -0.012 | 0.0425 | No | ||
| 92 | TRIM46 | 1460568_at | 14623 | -0.069 | 0.0219 | No | ||
| 93 | BCL9L | 1419180_at 1438806_at 1443700_at 1444855_at | 15693 | -0.218 | -0.0258 | No | ||
| 94 | ESRRA | 1442864_at 1460652_at | 16134 | -0.297 | -0.0443 | No | ||
| 95 | RLF | 1427171_at 1439555_at 1440020_at 1446057_at 1447164_at | 16268 | -0.323 | -0.0485 | No | ||
| 96 | B3GALT6 | 1421155_at 1435252_at 1440340_at | 16311 | -0.331 | -0.0485 | No | ||
| 97 | HNRPF | 1450963_at | 16388 | -0.346 | -0.0500 | No | ||
| 98 | ALDH3B1 | 1452301_at | 16477 | -0.364 | -0.0519 | No | ||
| 99 | CCDC6 | 1428311_at 1459159_a_at | 16680 | -0.414 | -0.0587 | No | ||
| 100 | FXYD2 | 1419378_a_at 1419379_x_at 1449883_at 1456601_x_at | 16710 | -0.424 | -0.0576 | No | ||
| 101 | CBX5 | 1421932_at 1421933_at 1421934_at 1450416_at 1454636_at 1459845_at | 16714 | -0.426 | -0.0552 | No | ||
| 102 | IL15RA | 1422397_a_at 1448681_at | 16780 | -0.440 | -0.0556 | No | ||
| 103 | ILK | 1449942_a_at | 17410 | -0.609 | -0.0809 | No | ||
| 104 | IVNS1ABP | 1420961_a_at 1425718_a_at 1450084_s_at | 17445 | -0.621 | -0.0789 | No | ||
| 105 | ADSS | 1460726_at | 17849 | -0.754 | -0.0930 | No | ||
| 106 | SC5DL | 1424709_at 1434520_at 1451457_at | 17859 | -0.758 | -0.0890 | No | ||
| 107 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 18047 | -0.827 | -0.0927 | No | ||
| 108 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 18107 | -0.846 | -0.0905 | No | ||
| 109 | NRAS | 1422688_a_at 1454060_a_at | 18126 | -0.853 | -0.0864 | No | ||
| 110 | AP3M1 | 1416375_at 1448308_at 1448309_at 1458092_at | 18141 | -0.858 | -0.0820 | No | ||
| 111 | ARHGAP20 | 1427522_at 1429918_at | 18146 | -0.861 | -0.0772 | No | ||
| 112 | MYCL1 | 1422087_at 1422088_at 1434777_at | 18223 | -0.888 | -0.0755 | No | ||
| 113 | HPS5 | 1434677_at | 18399 | -0.944 | -0.0780 | No | ||
| 114 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 18887 | -1.114 | -0.0939 | No | ||
| 115 | ERCC6 | 1441595_at 1442604_at | 19037 | -1.184 | -0.0938 | No | ||
| 116 | POGK | 1447864_s_at 1459896_at | 19099 | -1.213 | -0.0895 | No | ||
| 117 | FOXJ3 | 1435362_at 1438220_at 1459055_at | 19123 | -1.229 | -0.0834 | No | ||
| 118 | FXYD6 | 1417343_at | 19322 | -1.334 | -0.0847 | No | ||
| 119 | RPL22 | 1448398_s_at 1453118_s_at | 19346 | -1.346 | -0.0779 | No | ||
| 120 | H3F3A | 1423263_at 1442087_at | 19390 | -1.370 | -0.0719 | No | ||
| 121 | AMPD2 | 1426757_at 1438941_x_at | 19726 | -1.588 | -0.0781 | No | ||
| 122 | NIT1 | 1417468_at 1417469_at | 20303 | -2.026 | -0.0927 | No | ||
| 123 | RRAGC | 1415749_a_at | 20321 | -2.041 | -0.0816 | No | ||
| 124 | SHMT2 | 1426423_at 1434204_x_at 1455084_x_at 1455985_x_at | 20408 | -2.106 | -0.0732 | No | ||
| 125 | EIF3S10 | 1416659_at 1416660_at 1416661_at 1448425_at | 20531 | -2.241 | -0.0658 | No | ||
| 126 | ZFP91 | 1426326_at 1429615_at 1429616_at 1459444_at | 20668 | -2.409 | -0.0580 | No | ||
| 127 | TMEM24 | 1428095_a_at 1457585_at | 20723 | -2.481 | -0.0460 | No | ||
| 128 | CSDA | 1435800_a_at 1451012_a_at | 20792 | -2.563 | -0.0342 | No | ||
| 129 | UVRAG | 1454706_at | 20844 | -2.645 | -0.0211 | No | ||
| 130 | RFX5 | 1423103_at | 20991 | -2.890 | -0.0110 | No | ||
| 131 | SLC43A1 | 1440007_at 1453255_at | 21326 | -3.716 | -0.0047 | No | ||
| 132 | RORC | 1425792_a_at 1425793_a_at | 21670 | -5.593 | 0.0122 | No |