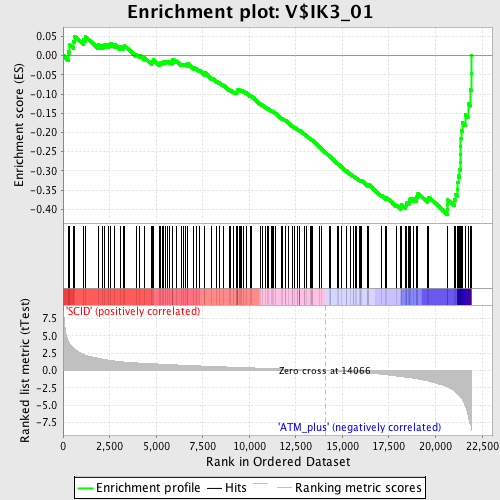
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$IK3_01 |
| Enrichment Score (ES) | -0.41358545 |
| Normalized Enrichment Score (NES) | -1.629257 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.10116416 |
| FWER p-Value | 0.467 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP1CB | 1426205_at 1431328_at 1433540_x_at 1456462_x_at | 287 | 4.199 | 0.0099 | No | ||
| 2 | MAP1LC3A | 1447883_x_at 1451290_at | 360 | 3.857 | 0.0278 | No | ||
| 3 | SLC6A9 | 1417636_at 1431812_a_at | 564 | 3.268 | 0.0365 | No | ||
| 4 | RCOR2 | 1417302_at | 627 | 3.122 | 0.0508 | No | ||
| 5 | PPAP2B | 1429514_at 1446850_at 1448908_at | 1106 | 2.314 | 0.0416 | No | ||
| 6 | CYP17A1 | 1417017_at | 1198 | 2.210 | 0.0496 | No | ||
| 7 | CHST8 | 1428977_at | 1883 | 1.791 | 0.0280 | No | ||
| 8 | USH3A | 1438347_at | 2105 | 1.668 | 0.0271 | No | ||
| 9 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 2241 | 1.602 | 0.0297 | No | ||
| 10 | HOXB4 | 1451761_at 1460379_at | 2455 | 1.499 | 0.0281 | No | ||
| 11 | STARD8 | 1427072_at | 2561 | 1.449 | 0.0313 | No | ||
| 12 | GNAO1 | 1421152_a_at 1448031_at 1460383_at | 2782 | 1.378 | 0.0288 | No | ||
| 13 | CCDC3 | 1428549_at | 3060 | 1.293 | 0.0232 | No | ||
| 14 | TNFRSF21 | 1422740_at 1450731_s_at | 3218 | 1.251 | 0.0228 | No | ||
| 15 | JAG1 | 1421105_at 1421106_at 1434070_at | 3277 | 1.238 | 0.0270 | No | ||
| 16 | CYR61 | 1416039_x_at 1417848_at 1417849_at 1438133_a_at 1442340_x_at 1457823_at 1459462_at | 3931 | 1.109 | 0.0031 | No | ||
| 17 | ASCL3 | 1420780_at | 4095 | 1.078 | 0.0016 | No | ||
| 18 | MYT1 | 1422773_at 1439365_at | 4382 | 1.035 | -0.0059 | No | ||
| 19 | RHBDF1 | 1424138_at | 4742 | 0.997 | -0.0169 | No | ||
| 20 | SNX16 | 1424276_at 1438803_s_at 1456041_at | 4807 | 0.989 | -0.0144 | No | ||
| 21 | CNTN4 | 1438782_at 1460321_at | 4837 | 0.985 | -0.0103 | No | ||
| 22 | MECP2 | 1438930_s_at 1449266_at 1460246_at | 5194 | 0.938 | -0.0215 | No | ||
| 23 | FLNC | 1424063_at 1447812_x_at 1449073_at | 5248 | 0.931 | -0.0188 | No | ||
| 24 | C2 | 1416051_at 1441912_x_at 1457664_x_at | 5321 | 0.921 | -0.0170 | No | ||
| 25 | NSD1 | 1420881_at 1435088_at 1440972_at 1456916_at | 5394 | 0.909 | -0.0153 | No | ||
| 26 | KCNA4 | 1422255_at 1438613_at | 5496 | 0.896 | -0.0150 | No | ||
| 27 | PTGES | 1439747_at 1449449_at 1449450_at | 5582 | 0.885 | -0.0141 | No | ||
| 28 | LMX1A | 1421554_at | 5713 | 0.867 | -0.0153 | No | ||
| 29 | FGF20 | 1421677_at | 5858 | 0.848 | -0.0172 | No | ||
| 30 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 5880 | 0.845 | -0.0135 | No | ||
| 31 | DHRS4 | 1419382_a_at 1451559_a_at | 5895 | 0.844 | -0.0095 | No | ||
| 32 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 6090 | 0.821 | -0.0139 | No | ||
| 33 | CDK9 | 1417269_at 1457240_at | 6358 | 0.786 | -0.0219 | No | ||
| 34 | TYRO3 | 1425248_a_at 1425249_a_at | 6450 | 0.774 | -0.0218 | No | ||
| 35 | TIMP3 | 1419088_at 1419089_at 1449334_at 1449335_at | 6547 | 0.762 | -0.0220 | No | ||
| 36 | SH3BGRL2 | 1434109_at 1434211_at | 6689 | 0.745 | -0.0244 | No | ||
| 37 | GABBR1 | 1422051_a_at 1425595_at 1437188_at 1455021_at | 6698 | 0.744 | -0.0207 | No | ||
| 38 | SYNPR | 1423640_at 1444331_at | 7011 | 0.709 | -0.0311 | No | ||
| 39 | SLITRK1 | 1428089_at | 7143 | 0.695 | -0.0333 | No | ||
| 40 | TMC4 | 1427178_at | 7336 | 0.671 | -0.0384 | No | ||
| 41 | PRRX1 | 1425526_a_at 1425527_at 1425528_at 1432129_a_at 1439774_at 1458004_at | 7611 | 0.640 | -0.0475 | No | ||
| 42 | NHLH2 | 1436888_at | 7617 | 0.639 | -0.0442 | No | ||
| 43 | RBX1 | 1416577_a_at 1416578_at 1438623_x_at 1440061_at 1447793_x_at 1448387_at | 7995 | 0.601 | -0.0582 | No | ||
| 44 | APBA1 | 1437940_at 1455369_at 1459605_at | 8262 | 0.574 | -0.0672 | No | ||
| 45 | TRAPPC3 | 1416771_at | 8388 | 0.561 | -0.0699 | No | ||
| 46 | AMBN | 1449417_at | 8596 | 0.540 | -0.0764 | No | ||
| 47 | SPINLW1 | 1420588_at | 8923 | 0.509 | -0.0886 | No | ||
| 48 | VAMP3 | 1421102_a_at 1433693_x_at 1433916_at 1437708_x_at 1456245_x_at 1457391_at | 9006 | 0.499 | -0.0896 | No | ||
| 49 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 9174 | 0.482 | -0.0946 | No | ||
| 50 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 9286 | 0.473 | -0.0971 | No | ||
| 51 | KIFC3 | 1416199_at | 9331 | 0.468 | -0.0966 | No | ||
| 52 | SLC30A3 | 1460654_at | 9348 | 0.466 | -0.0947 | No | ||
| 53 | HMGA2 | 1422851_at 1443394_at 1446196_at 1450780_s_at 1450781_at 1458241_at | 9358 | 0.465 | -0.0926 | No | ||
| 54 | WNT3 | 1450763_x_at | 9366 | 0.464 | -0.0904 | No | ||
| 55 | NEDD1 | 1423196_at 1450899_at | 9389 | 0.463 | -0.0888 | No | ||
| 56 | SLC26A6 | 1416275_at | 9451 | 0.456 | -0.0891 | No | ||
| 57 | T | 1419304_at | 9520 | 0.449 | -0.0898 | No | ||
| 58 | ADAM15 | 1416080_at 1425170_a_at 1438760_x_at 1454206_a_at | 9586 | 0.442 | -0.0903 | No | ||
| 59 | CD3E | 1422105_at 1445748_at | 9711 | 0.429 | -0.0937 | No | ||
| 60 | MKNK2 | 1418300_a_at 1449029_at | 9830 | 0.419 | -0.0968 | No | ||
| 61 | UPP2 | 1460059_at | 10049 | 0.396 | -0.1046 | No | ||
| 62 | SMPX | 1418095_at | 10121 | 0.390 | -0.1057 | No | ||
| 63 | RGS5 | 1420940_x_at 1420941_at | 10579 | 0.347 | -0.1248 | No | ||
| 64 | SEMA3A | 1420416_at 1420417_at 1449865_at | 10699 | 0.336 | -0.1284 | No | ||
| 65 | FGF10 | 1420690_at | 10875 | 0.318 | -0.1347 | No | ||
| 66 | C1S | 1424041_s_at | 10977 | 0.310 | -0.1376 | No | ||
| 67 | RAMP2 | 1418187_at | 11041 | 0.305 | -0.1388 | No | ||
| 68 | PPP2R2B | 1426621_a_at | 11190 | 0.293 | -0.1440 | No | ||
| 69 | ANK3 | 1425202_a_at 1439220_at 1447259_at 1451628_a_at 1457288_at | 11235 | 0.288 | -0.1445 | No | ||
| 70 | SIX5 | 1427560_at | 11297 | 0.281 | -0.1457 | No | ||
| 71 | SLC26A7 | 1425841_at | 11426 | 0.268 | -0.1501 | No | ||
| 72 | HOXB9 | 1452317_at | 11753 | 0.233 | -0.1638 | No | ||
| 73 | VSNL1 | 1420955_at 1450055_at | 11806 | 0.228 | -0.1649 | No | ||
| 74 | BAI3 | 1446358_at 1454782_at | 11916 | 0.216 | -0.1688 | No | ||
| 75 | SLIT3 | 1427086_at 1452296_at | 11920 | 0.216 | -0.1677 | No | ||
| 76 | TAL1 | 1449389_at | 12092 | 0.200 | -0.1745 | No | ||
| 77 | SERPINI1 | 1416702_at 1443756_at 1448443_at | 12323 | 0.180 | -0.1840 | No | ||
| 78 | MAP4K4 | 1422615_at 1440609_at 1448050_s_at 1459912_at | 12344 | 0.179 | -0.1840 | No | ||
| 79 | NNMT | 1432517_a_at | 12446 | 0.169 | -0.1877 | No | ||
| 80 | MAG | 1460219_at | 12566 | 0.158 | -0.1923 | No | ||
| 81 | IGFALS | 1422826_at | 12598 | 0.155 | -0.1928 | No | ||
| 82 | NAPG | 1424069_at 1424070_at 1443366_at | 12677 | 0.148 | -0.1956 | No | ||
| 83 | LIFR | 1425107_a_at 1450207_at | 12686 | 0.147 | -0.1952 | No | ||
| 84 | OTX1 | 1437601_at | 12687 | 0.147 | -0.1944 | No | ||
| 85 | CHAD | 1420569_at | 12698 | 0.146 | -0.1940 | No | ||
| 86 | DNAH9 | 1442412_at 1457401_at | 12721 | 0.144 | -0.1942 | No | ||
| 87 | DIO2 | 1418937_at 1418938_at 1426081_a_at | 12980 | 0.118 | -0.2054 | No | ||
| 88 | LHX9 | 1419324_at 1431598_a_at 1441313_x_at | 12981 | 0.118 | -0.2048 | No | ||
| 89 | HCST | 1419119_at | 13083 | 0.107 | -0.2088 | No | ||
| 90 | BNC2 | 1438861_at 1440593_at 1459132_at | 13306 | 0.081 | -0.2186 | No | ||
| 91 | LMO6 | 1439296_at | 13339 | 0.077 | -0.2196 | No | ||
| 92 | DNAJB8 | 1449223_at | 13354 | 0.076 | -0.2198 | No | ||
| 93 | PSD | 1435780_at | 13404 | 0.071 | -0.2217 | No | ||
| 94 | CCL20 | 1422029_at | 13409 | 0.070 | -0.2215 | No | ||
| 95 | PCDH10 | 1421311_at 1425563_s_at 1430667_at 1438134_at 1442565_at 1444301_at | 13755 | 0.033 | -0.2372 | No | ||
| 96 | CXCL5 | 1419728_at | 13857 | 0.022 | -0.2417 | No | ||
| 97 | SCML4 | 1427417_at 1427545_at 1440157_at | 14310 | -0.029 | -0.2623 | No | ||
| 98 | ACCN2 | 1455328_at | 14347 | -0.033 | -0.2637 | No | ||
| 99 | PROK2 | 1419681_a_at 1426176_a_at 1451952_at | 14369 | -0.036 | -0.2645 | No | ||
| 100 | SALL1 | 1450489_at | 14738 | -0.085 | -0.2809 | No | ||
| 101 | GRIN2B | 1422223_at 1431700_at | 14788 | -0.090 | -0.2827 | No | ||
| 102 | HECTD1 | 1424141_at 1438039_at | 14976 | -0.112 | -0.2907 | No | ||
| 103 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 15205 | -0.145 | -0.3003 | No | ||
| 104 | ACE2 | 1425102_a_at 1425103_at 1452138_a_at | 15214 | -0.145 | -0.2999 | No | ||
| 105 | SLC38A5 | 1454622_at | 15416 | -0.178 | -0.3082 | No | ||
| 106 | RTN1 | 1429761_at | 15587 | -0.202 | -0.3148 | No | ||
| 107 | AMMECR1 | 1430697_at 1450546_at | 15611 | -0.206 | -0.3148 | No | ||
| 108 | SFRS16 | 1428961_a_at | 15728 | -0.221 | -0.3189 | No | ||
| 109 | RBBP9 | 1416174_at | 15761 | -0.228 | -0.3191 | No | ||
| 110 | STARD3 | 1417405_at | 15896 | -0.253 | -0.3239 | No | ||
| 111 | PUNC | 1423496_a_at 1439498_at 1444981_at | 15981 | -0.269 | -0.3262 | No | ||
| 112 | CEPT1 | 1416471_at 1443090_at 1446410_at 1459406_at | 15992 | -0.270 | -0.3252 | No | ||
| 113 | AP1G2 | 1419113_at | 16047 | -0.281 | -0.3261 | No | ||
| 114 | FGF17 | 1421523_at 1456239_at | 16350 | -0.338 | -0.3381 | No | ||
| 115 | USP34 | 1434392_at 1434393_at 1444437_at 1446112_at 1460126_at | 16370 | -0.343 | -0.3371 | No | ||
| 116 | XPO1 | 1418442_at 1418443_at 1448070_at | 16374 | -0.344 | -0.3354 | No | ||
| 117 | E2F4 | 1451480_at | 16422 | -0.353 | -0.3356 | No | ||
| 118 | CBX8 | 1419198_at 1419199_at | 17086 | -0.518 | -0.3632 | No | ||
| 119 | MSX1 | 1417127_at 1448601_s_at | 17320 | -0.585 | -0.3707 | No | ||
| 120 | PKP4 | 1438677_at 1452209_at | 17374 | -0.601 | -0.3698 | No | ||
| 121 | TRPM3 | 1439026_at 1441966_at 1443723_at 1445555_at 1456633_at 1456923_at 1457927_at | 17898 | -0.770 | -0.3896 | No | ||
| 122 | HCN3 | 1451023_at | 18115 | -0.847 | -0.3948 | No | ||
| 123 | POU4F2 | 1437588_at | 18173 | -0.869 | -0.3927 | No | ||
| 124 | CHRNB2 | 1420744_at 1436428_at 1441837_at | 18177 | -0.872 | -0.3880 | No | ||
| 125 | BAZ2B | 1435240_at 1440984_at 1441556_at 1442704_at 1442971_at 1445048_at 1448007_at 1458077_at | 18390 | -0.942 | -0.3926 | No | ||
| 126 | SIRT6 | 1429631_at | 18418 | -0.951 | -0.3886 | No | ||
| 127 | TGIF2 | 1431115_at 1434400_at | 18425 | -0.955 | -0.3836 | No | ||
| 128 | USP25 | 1442104_at 1442826_at 1445116_at 1445861_at 1448939_at | 18539 | -0.992 | -0.3833 | No | ||
| 129 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 18594 | -1.004 | -0.3803 | No | ||
| 130 | MKL1 | 1434900_at | 18597 | -1.005 | -0.3748 | No | ||
| 131 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 18671 | -1.030 | -0.3725 | No | ||
| 132 | RHOQ | 1427918_a_at 1440925_at | 18807 | -1.081 | -0.3728 | No | ||
| 133 | POLR2B | 1433552_a_at | 18979 | -1.164 | -0.3742 | No | ||
| 134 | TNNI2 | 1416889_at 1438608_at 1438609_x_at | 18981 | -1.164 | -0.3679 | No | ||
| 135 | USP52 | 1426700_a_at | 19020 | -1.177 | -0.3631 | No | ||
| 136 | TCF8 | 1418926_at 1420243_at 1443005_at 1443203_at 1446127_at 1456686_at | 19055 | -1.192 | -0.3581 | No | ||
| 137 | MBNL2 | 1433754_at 1436858_at 1446475_at 1455827_at 1457237_at 1457477_at | 19582 | -1.491 | -0.3741 | No | ||
| 138 | C7ORF10 | 1424597_at 1451404_at | 19641 | -1.532 | -0.3683 | No | ||
| 139 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 20628 | -2.369 | -0.4006 | Yes | ||
| 140 | MTSS1 | 1424826_s_at 1434036_at 1440847_at 1443729_at 1446284_at 1451496_at | 20660 | -2.399 | -0.3888 | Yes | ||
| 141 | OSBPL7 | 1428893_at 1459966_at | 20664 | -2.402 | -0.3757 | Yes | ||
| 142 | MADD | 1425735_at 1455502_at | 21006 | -2.926 | -0.3753 | Yes | ||
| 143 | STAG2 | 1421849_at 1450396_at | 21067 | -3.048 | -0.3612 | Yes | ||
| 144 | RAB11B | 1423448_at | 21163 | -3.275 | -0.3476 | Yes | ||
| 145 | KCNH2 | 1449544_a_at | 21191 | -3.341 | -0.3305 | Yes | ||
| 146 | ARPC2 | 1437148_at 1442295_at | 21214 | -3.402 | -0.3127 | Yes | ||
| 147 | FOS | 1423100_at | 21307 | -3.658 | -0.2968 | Yes | ||
| 148 | GATA3 | 1448886_at | 21331 | -3.739 | -0.2773 | Yes | ||
| 149 | SPIB | 1460407_at | 21336 | -3.766 | -0.2568 | Yes | ||
| 150 | EIF4A2 | 1450934_at | 21350 | -3.811 | -0.2364 | Yes | ||
| 151 | HIVEP1 | 1422742_at | 21365 | -3.861 | -0.2158 | Yes | ||
| 152 | UBXD2 | 1426484_at 1426485_at 1426486_at | 21393 | -3.955 | -0.1953 | Yes | ||
| 153 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 21442 | -4.147 | -0.1747 | Yes | ||
| 154 | EDEM1 | 1424065_at 1451218_at | 21594 | -5.067 | -0.1538 | Yes | ||
| 155 | WSB2 | 1421846_at 1421847_at 1438129_at | 21747 | -6.370 | -0.1257 | Yes | ||
| 156 | PDCD10 | 1437530_at 1448527_at 1448528_at | 21867 | -7.732 | -0.0886 | Yes | ||
| 157 | EGR3 | 1421486_at 1436329_at | 21923 | -8.252 | -0.0457 | Yes | ||
| 158 | EGR1 | 1417065_at | 21931 | -8.413 | 0.0002 | Yes |