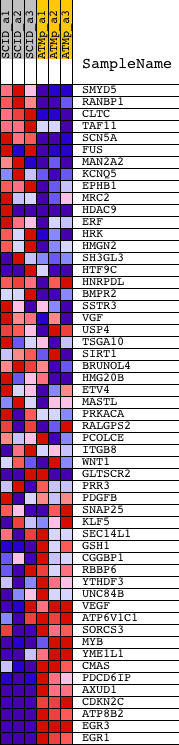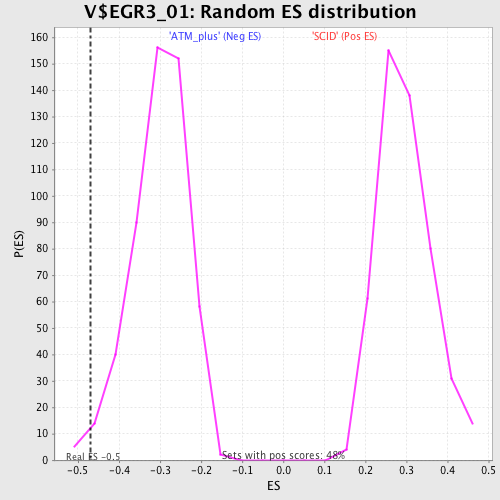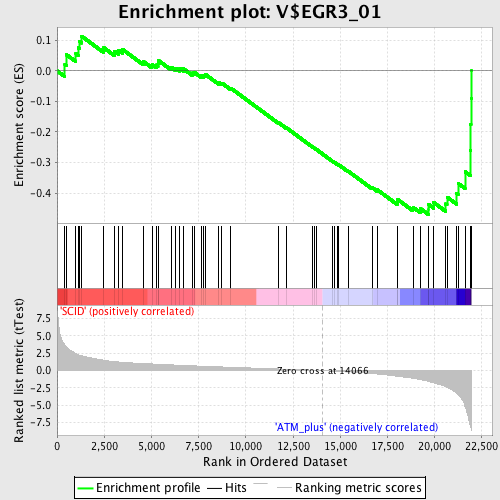
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$EGR3_01 |
| Enrichment Score (ES) | -0.47017333 |
| Normalized Enrichment Score (NES) | -1.5597204 |
| Nominal p-value | 0.021276595 |
| FDR q-value | 0.12758227 |
| FWER p-Value | 0.771 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SMYD5 | 1425479_at 1447749_at 1451722_s_at | 412 | 3.679 | 0.0203 | No | ||
| 2 | RANBP1 | 1422547_at | 490 | 3.453 | 0.0534 | No | ||
| 3 | CLTC | 1419407_at 1439094_at 1440457_at 1454626_at | 994 | 2.455 | 0.0565 | No | ||
| 4 | TAF11 | 1446161_at 1451995_at | 1115 | 2.306 | 0.0755 | No | ||
| 5 | SCN5A | 1422194_at 1458813_at | 1177 | 2.229 | 0.0964 | No | ||
| 6 | FUS | 1451285_at 1451286_s_at 1455831_at 1458801_at | 1291 | 2.137 | 0.1139 | No | ||
| 7 | MAN2A2 | 1435203_at 1457498_at | 2473 | 1.489 | 0.0758 | No | ||
| 8 | KCNQ5 | 1427426_at 1441071_at | 3034 | 1.302 | 0.0640 | No | ||
| 9 | EPHB1 | 1455188_at | 3253 | 1.242 | 0.0672 | No | ||
| 10 | MRC2 | 1421044_at 1421045_at 1446466_at | 3460 | 1.198 | 0.0705 | No | ||
| 11 | HDAC9 | 1421306_a_at 1434572_at | 4559 | 1.014 | 0.0311 | No | ||
| 12 | ERF | 1422114_at 1435561_at | 5026 | 0.962 | 0.0201 | No | ||
| 13 | HRK | 1439854_at 1450366_at | 5257 | 0.931 | 0.0194 | No | ||
| 14 | HMGN2 | 1433507_a_at | 5345 | 0.917 | 0.0252 | No | ||
| 15 | SH3GL3 | 1432103_a_at | 5361 | 0.914 | 0.0342 | No | ||
| 16 | HTF9C | 1434244_x_at 1441920_x_at 1448114_a_at 1448115_at 1453995_a_at 1457620_at | 6042 | 0.827 | 0.0119 | No | ||
| 17 | HNRPDL | 1420093_s_at 1420094_at 1424251_a_at 1424252_at 1428224_at 1428225_s_at 1449039_a_at 1456698_s_at | 6283 | 0.795 | 0.0094 | No | ||
| 18 | BMPR2 | 1419616_at 1434310_at 1441652_at | 6503 | 0.766 | 0.0075 | No | ||
| 19 | SSTR3 | 1441603_at 1450577_at | 6690 | 0.745 | 0.0070 | No | ||
| 20 | VGF | 1436094_at | 7152 | 0.693 | -0.0068 | No | ||
| 21 | USP4 | 1442319_at 1450892_a_at | 7286 | 0.677 | -0.0056 | No | ||
| 22 | TSGA10 | 1436045_at 1447283_at | 7629 | 0.638 | -0.0145 | No | ||
| 23 | SIRT1 | 1418640_at 1458538_at | 7772 | 0.623 | -0.0144 | No | ||
| 24 | BRUNOL4 | 1426929_at 1426930_at 1452240_at 1458475_at | 7858 | 0.615 | -0.0117 | No | ||
| 25 | HMG20B | 1417637_a_at | 8569 | 0.542 | -0.0384 | No | ||
| 26 | ETV4 | 1423232_at 1443381_at | 8726 | 0.527 | -0.0399 | No | ||
| 27 | MASTL | 1423524_at 1423525_at 1444725_at 1447136_at | 9207 | 0.479 | -0.0568 | No | ||
| 28 | PRKACA | 1447720_x_at 1450519_a_at | 11710 | 0.238 | -0.1686 | No | ||
| 29 | RALGPS2 | 1417230_at 1428789_at 1431704_a_at | 12143 | 0.196 | -0.1862 | No | ||
| 30 | PCOLCE | 1437165_a_at 1448433_a_at | 13517 | 0.058 | -0.2484 | No | ||
| 31 | ITGB8 | 1436223_at | 13641 | 0.045 | -0.2535 | No | ||
| 32 | WNT1 | 1425377_at | 13733 | 0.035 | -0.2573 | No | ||
| 33 | GLTSCR2 | 1423803_s_at 1451121_a_at 1452409_at | 14588 | -0.064 | -0.2957 | No | ||
| 34 | PRR3 | 1424145_at | 14695 | -0.078 | -0.2997 | No | ||
| 35 | PDGFB | 1450413_at 1450414_at | 14850 | -0.097 | -0.3057 | No | ||
| 36 | SNAP25 | 1416828_at 1443398_at | 14921 | -0.106 | -0.3078 | No | ||
| 37 | KLF5 | 1430255_at 1451021_a_at 1451739_at | 15413 | -0.177 | -0.3283 | No | ||
| 38 | SEC14L1 | 1451908_a_at 1453412_a_at | 16677 | -0.413 | -0.3817 | No | ||
| 39 | GSH1 | 1450002_at | 16949 | -0.484 | -0.3889 | No | ||
| 40 | CGGBP1 | 1454641_at 1455658_at | 18011 | -0.809 | -0.4288 | No | ||
| 41 | RBBP6 | 1425114_at 1425115_at 1425421_at 1426487_a_at 1453611_at | 18045 | -0.825 | -0.4215 | No | ||
| 42 | YTHDF3 | 1426840_at 1426841_at 1426842_at 1445235_at | 18849 | -1.099 | -0.4466 | No | ||
| 43 | UNC84B | 1433832_at 1439238_at | 19257 | -1.296 | -0.4514 | No | ||
| 44 | VEGF | 1420909_at 1451959_a_at | 19669 | -1.555 | -0.4537 | Yes | ||
| 45 | ATP6V1C1 | 1419544_at 1419545_a_at 1419546_at | 19672 | -1.555 | -0.4372 | Yes | ||
| 46 | SORCS3 | 1425110_at 1425111_at | 19943 | -1.758 | -0.4309 | Yes | ||
| 47 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 20588 | -2.317 | -0.4357 | Yes | ||
| 48 | YME1L1 | 1423360_at 1423361_at 1437822_at 1450954_at | 20652 | -2.394 | -0.4132 | Yes | ||
| 49 | CMAS | 1426662_at | 21162 | -3.275 | -0.4017 | Yes | ||
| 50 | PDCD6IP | 1415937_s_at 1426184_a_at 1448155_at 1449674_s_at 1460263_at | 21240 | -3.469 | -0.3683 | Yes | ||
| 51 | AXUD1 | 1434350_at | 21608 | -5.152 | -0.3304 | Yes | ||
| 52 | CDKN2C | 1416868_at 1439164_at | 21886 | -7.816 | -0.2600 | Yes | ||
| 53 | ATP8B2 | 1434026_at 1441699_at 1452450_at | 21910 | -8.016 | -0.1759 | Yes | ||
| 54 | EGR3 | 1421486_at 1436329_at | 21923 | -8.252 | -0.0888 | Yes | ||
| 55 | EGR1 | 1417065_at | 21931 | -8.413 | 0.0002 | Yes |