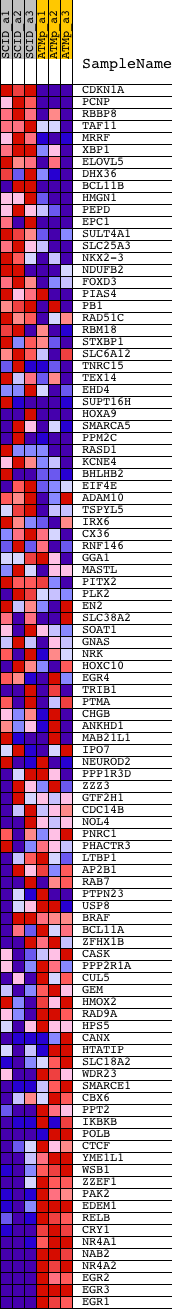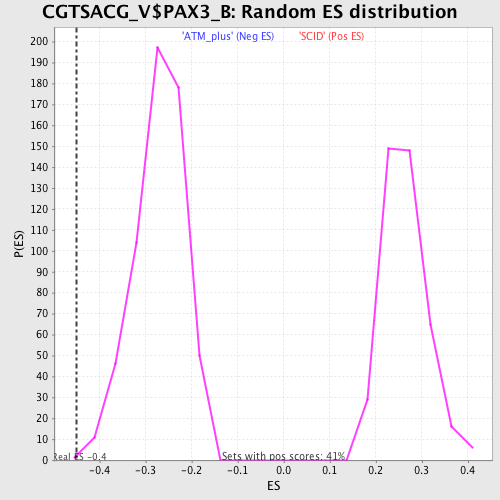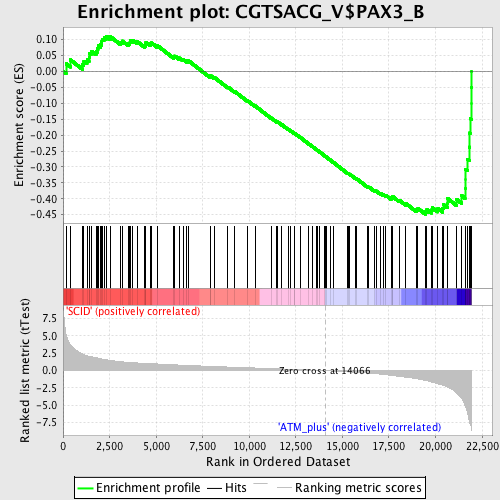
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CGTSACG_V$PAX3_B |
| Enrichment Score (ES) | -0.4493487 |
| Normalized Enrichment Score (NES) | -1.6687561 |
| Nominal p-value | 0.0017035775 |
| FDR q-value | 0.10221206 |
| FWER p-Value | 0.325 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDKN1A | 1421679_a_at 1424638_at | 161 | 5.189 | 0.0241 | No | ||
| 2 | PCNP | 1428314_at 1452735_at | 386 | 3.753 | 0.0366 | No | ||
| 3 | RBBP8 | 1427061_at 1427062_at 1446097_at | 1039 | 2.403 | 0.0213 | No | ||
| 4 | TAF11 | 1446161_at 1451995_at | 1115 | 2.306 | 0.0318 | No | ||
| 5 | MRRF | 1426364_at | 1284 | 2.140 | 0.0371 | No | ||
| 6 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 1394 | 2.059 | 0.0446 | No | ||
| 7 | ELOVL5 | 1415840_at 1437211_x_at 1459242_at | 1419 | 2.044 | 0.0558 | No | ||
| 8 | DHX36 | 1424397_at 1424398_at 1437550_at | 1503 | 2.001 | 0.0642 | No | ||
| 9 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 1772 | 1.860 | 0.0632 | No | ||
| 10 | HMGN1 | 1422495_a_at 1422496_at 1438940_x_at 1455897_x_at | 1827 | 1.828 | 0.0718 | No | ||
| 11 | PEPD | 1416712_at | 1875 | 1.796 | 0.0805 | No | ||
| 12 | EPC1 | 1418850_at 1429914_at 1440198_at 1440742_at 1442279_at 1443675_at 1458315_at | 2019 | 1.711 | 0.0843 | No | ||
| 13 | SULT4A1 | 1421606_a_at 1433714_at | 2058 | 1.694 | 0.0928 | No | ||
| 14 | SLC25A3 | 1416300_a_at 1440302_at | 2088 | 1.675 | 0.1017 | No | ||
| 15 | NKX2-3 | 1425827_at 1427741_x_at 1452528_a_at | 2199 | 1.619 | 0.1064 | No | ||
| 16 | NDUFB2 | 1416834_x_at 1442658_at 1448483_a_at 1453806_at | 2308 | 1.563 | 0.1110 | No | ||
| 17 | FOXD3 | 1422210_at | 2547 | 1.454 | 0.1089 | No | ||
| 18 | PIAS4 | 1418861_at 1455394_at | 3080 | 1.289 | 0.0923 | No | ||
| 19 | PB1 | 1426877_a_at 1426878_at 1427266_at 1442603_at | 3174 | 1.261 | 0.0957 | No | ||
| 20 | RAD51C | 1423026_at 1438453_at | 3501 | 1.187 | 0.0880 | No | ||
| 21 | RBM18 | 1420607_at 1420608_at 1458182_at | 3590 | 1.168 | 0.0910 | No | ||
| 22 | STXBP1 | 1420505_a_at 1420506_a_at | 3613 | 1.163 | 0.0971 | No | ||
| 23 | SLC6A12 | 1449382_at | 3751 | 1.140 | 0.0977 | No | ||
| 24 | TNRC15 | 1439837_at 1446275_at 1451856_at 1457472_at | 3984 | 1.097 | 0.0937 | No | ||
| 25 | TEX14 | 1419240_at | 4373 | 1.036 | 0.0822 | No | ||
| 26 | EHD4 | 1449852_a_at | 4436 | 1.027 | 0.0856 | No | ||
| 27 | SUPT16H | 1419741_at 1449578_at 1456449_at | 4446 | 1.026 | 0.0914 | No | ||
| 28 | HOXA9 | 1421579_at 1455626_at | 4676 | 1.002 | 0.0870 | No | ||
| 29 | SMARCA5 | 1440048_at | 4750 | 0.995 | 0.0897 | No | ||
| 30 | PPM2C | 1434228_at 1438201_at | 5054 | 0.959 | 0.0816 | No | ||
| 31 | RASD1 | 1423619_at | 5939 | 0.838 | 0.0462 | No | ||
| 32 | KCNE4 | 1418156_at | 5999 | 0.832 | 0.0486 | No | ||
| 33 | BHLHB2 | 1418025_at | 6240 | 0.800 | 0.0424 | No | ||
| 34 | EIF4E | 1450909_at 1457489_at 1459371_at | 6449 | 0.774 | 0.0376 | No | ||
| 35 | ADAM10 | 1450104_at 1460083_at | 6631 | 0.753 | 0.0338 | No | ||
| 36 | TSPYL5 | 1455807_at | 6743 | 0.740 | 0.0332 | No | ||
| 37 | IRX6 | 1427383_at | 7918 | 0.609 | -0.0168 | No | ||
| 38 | CX36 | 1423019_at 1435297_at | 7924 | 0.608 | -0.0134 | No | ||
| 39 | RNF146 | 1417733_at 1425001_at 1425713_a_at | 8124 | 0.586 | -0.0189 | No | ||
| 40 | GGA1 | 1423834_s_at 1459863_x_at | 8848 | 0.516 | -0.0489 | No | ||
| 41 | MASTL | 1423524_at 1423525_at 1444725_at 1447136_at | 9207 | 0.479 | -0.0624 | No | ||
| 42 | PITX2 | 1424797_a_at 1450482_a_at | 9912 | 0.411 | -0.0922 | No | ||
| 43 | PLK2 | 1427005_at | 10313 | 0.373 | -0.1082 | No | ||
| 44 | EN2 | 1418868_at | 11180 | 0.293 | -0.1461 | No | ||
| 45 | SLC38A2 | 1426722_at 1429593_at 1429594_at | 11476 | 0.263 | -0.1580 | No | ||
| 46 | SOAT1 | 1417695_a_at 1417696_at 1417697_at 1426818_at 1448068_at | 11491 | 0.262 | -0.1571 | No | ||
| 47 | GNAS | 1421740_at 1427789_s_at 1443007_at 1443375_at 1444767_at 1450186_s_at 1453413_at | 11745 | 0.234 | -0.1673 | No | ||
| 48 | NRK | 1436399_s_at 1450078_at 1450079_at | 12087 | 0.201 | -0.1817 | No | ||
| 49 | HOXC10 | 1439798_at | 12216 | 0.190 | -0.1864 | No | ||
| 50 | EGR4 | 1449977_at | 12420 | 0.171 | -0.1946 | No | ||
| 51 | TRIB1 | 1424880_at 1424881_at | 12443 | 0.169 | -0.1946 | No | ||
| 52 | PTMA | 1423455_at | 12741 | 0.143 | -0.2073 | No | ||
| 53 | CHGB | 1415885_at | 13199 | 0.094 | -0.2277 | No | ||
| 54 | ANKHD1 | 1419867_a_at 1436597_at 1440754_at 1441597_at 1442641_at 1448008_at 1453023_at 1455759_a_at | 13400 | 0.071 | -0.2364 | No | ||
| 55 | MAB21L1 | 1421369_a_at 1424679_at 1440860_at 1457290_at | 13599 | 0.049 | -0.2452 | No | ||
| 56 | IPO7 | 1442511_at 1454955_at 1458165_at | 13657 | 0.044 | -0.2476 | No | ||
| 57 | NEUROD2 | 1444362_at | 13788 | 0.029 | -0.2533 | No | ||
| 58 | PPP1R3D | 1452922_at | 14035 | 0.003 | -0.2646 | No | ||
| 59 | ZZZ3 | 1434332_at 1438487_s_at 1441748_at | 14101 | -0.004 | -0.2675 | No | ||
| 60 | GTF2H1 | 1453169_a_at | 14164 | -0.012 | -0.2703 | No | ||
| 61 | CDC14B | 1429418_at 1437070_at | 14340 | -0.032 | -0.2781 | No | ||
| 62 | NOL4 | 1430189_at 1441447_at 1456387_at | 14509 | -0.055 | -0.2855 | No | ||
| 63 | PNRC1 | 1433668_at 1438524_x_at 1439513_at | 15263 | -0.153 | -0.3190 | No | ||
| 64 | PHACTR3 | 1429651_at | 15325 | -0.162 | -0.3209 | No | ||
| 65 | LTBP1 | 1419786_at 1440678_at 1446232_at 1447547_at 1448870_at 1457852_at 1458739_at | 15406 | -0.175 | -0.3235 | No | ||
| 66 | AP2B1 | 1427077_a_at 1445142_at 1446566_at 1452292_at 1458971_at | 15683 | -0.217 | -0.3348 | No | ||
| 67 | RAB7 | 1415734_at 1448159_at | 15765 | -0.228 | -0.3371 | No | ||
| 68 | PTPN23 | 1427128_at | 16343 | -0.337 | -0.3615 | No | ||
| 69 | USP8 | 1422883_at 1443420_at 1446807_at 1447386_at | 16426 | -0.353 | -0.3631 | No | ||
| 70 | BRAF | 1425693_at 1435434_at 1435480_at 1442749_at 1445786_at 1447940_a_at 1447941_x_at 1456505_at 1458641_at | 16748 | -0.432 | -0.3752 | No | ||
| 71 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 16815 | -0.447 | -0.3755 | No | ||
| 72 | ZFHX1B | 1422748_at 1434298_at 1442393_at 1445518_at 1454200_at 1456389_at | 17050 | -0.506 | -0.3832 | No | ||
| 73 | CASK | 1422518_at 1422519_at 1427692_a_at 1445152_at | 17210 | -0.551 | -0.3871 | No | ||
| 74 | PPP2R1A | 1415819_a_at 1438174_x_at 1438383_x_at 1438991_x_at 1455929_x_at | 17329 | -0.587 | -0.3890 | No | ||
| 75 | CUL5 | 1428287_at 1452722_a_at 1456102_a_at | 17616 | -0.679 | -0.3979 | No | ||
| 76 | GEM | 1426063_a_at | 17669 | -0.693 | -0.3961 | No | ||
| 77 | HMOX2 | 1416399_a_at | 17689 | -0.698 | -0.3928 | No | ||
| 78 | RAD9A | 1418404_at | 18055 | -0.831 | -0.4044 | No | ||
| 79 | HPS5 | 1434677_at | 18399 | -0.944 | -0.4144 | No | ||
| 80 | CANX | 1415692_s_at 1422845_at 1428935_at | 18968 | -1.156 | -0.4334 | No | ||
| 81 | HTATIP | 1433980_at 1433981_s_at | 19053 | -1.191 | -0.4301 | No | ||
| 82 | SLC18A2 | 1437079_at | 19475 | -1.425 | -0.4407 | Yes | ||
| 83 | WDR23 | 1448418_s_at 1460189_at | 19537 | -1.472 | -0.4346 | Yes | ||
| 84 | SMARCE1 | 1422675_at 1422676_at 1430822_at | 19764 | -1.620 | -0.4351 | Yes | ||
| 85 | CBX6 | 1424407_s_at 1429290_at | 19821 | -1.663 | -0.4276 | Yes | ||
| 86 | PPT2 | 1418302_at | 20127 | -1.907 | -0.4300 | Yes | ||
| 87 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 20388 | -2.086 | -0.4293 | Yes | ||
| 88 | POLB | 1425371_at 1434229_a_at | 20420 | -2.119 | -0.4179 | Yes | ||
| 89 | CTCF | 1418330_at 1449042_at | 20645 | -2.384 | -0.4137 | Yes | ||
| 90 | YME1L1 | 1423360_at 1423361_at 1437822_at 1450954_at | 20652 | -2.394 | -0.3994 | Yes | ||
| 91 | WSB1 | 1425241_a_at | 21123 | -3.169 | -0.4018 | Yes | ||
| 92 | ZZEF1 | 1434192_at 1438691_at 1446081_at 1453583_at | 21407 | -4.014 | -0.3904 | Yes | ||
| 93 | PAK2 | 1434250_at 1454887_at | 21587 | -4.997 | -0.3683 | Yes | ||
| 94 | EDEM1 | 1424065_at 1451218_at | 21594 | -5.067 | -0.3379 | Yes | ||
| 95 | RELB | 1417856_at | 21596 | -5.083 | -0.3071 | Yes | ||
| 96 | CRY1 | 1433733_a_at | 21708 | -5.979 | -0.2760 | Yes | ||
| 97 | NR4A1 | 1416505_at | 21809 | -7.139 | -0.2373 | Yes | ||
| 98 | NAB2 | 1417930_at | 21843 | -7.510 | -0.1933 | Yes | ||
| 99 | NR4A2 | 1447863_s_at 1450749_a_at 1450750_a_at 1455034_at | 21861 | -7.700 | -0.1474 | Yes | ||
| 100 | EGR2 | 1427682_a_at 1427683_at | 21921 | -8.208 | -0.1004 | Yes | ||
| 101 | EGR3 | 1421486_at 1436329_at | 21923 | -8.252 | -0.0504 | Yes | ||
| 102 | EGR1 | 1417065_at | 21931 | -8.413 | 0.0002 | Yes |