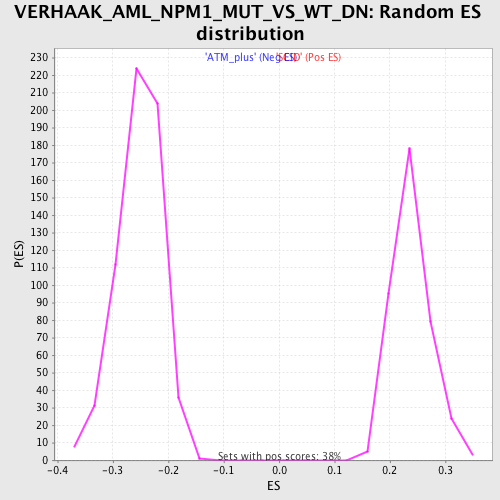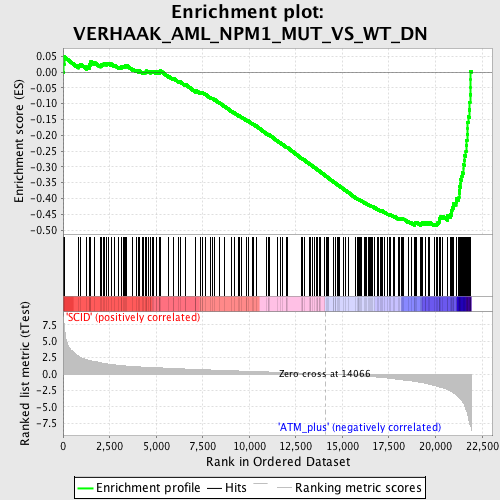
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | VERHAAK_AML_NPM1_MUT_VS_WT_DN |
| Enrichment Score (ES) | -0.4870413 |
| Normalized Enrichment Score (NES) | -1.9322357 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.012070375 |
| FWER p-Value | 0.184 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ASS1 | 1416239_at 1459937_at | 22 | 7.949 | 0.0249 | No | ||
| 2 | TSPAN7 | 1417502_at 1448737_at 1459812_x_at | 52 | 7.558 | 0.0481 | No | ||
| 3 | KIT | 1415900_a_at 1452514_a_at 1459588_at | 820 | 2.727 | 0.0217 | No | ||
| 4 | GALC | 1420547_at 1449900_at 1452907_at | 938 | 2.531 | 0.0246 | No | ||
| 5 | CST7 | 1419202_at 1445263_at | 1271 | 2.150 | 0.0163 | No | ||
| 6 | MX1 | 1451905_a_at | 1408 | 2.049 | 0.0167 | No | ||
| 7 | ATP9A | 1415932_x_at 1425838_at 1427604_a_at | 1411 | 2.049 | 0.0233 | No | ||
| 8 | HSPG2 | 1418669_at 1418670_s_at | 1454 | 2.022 | 0.0279 | No | ||
| 9 | POLE | 1441854_at 1448650_a_at | 1491 | 2.004 | 0.0328 | No | ||
| 10 | GNG7 | 1455190_at | 1699 | 1.901 | 0.0295 | No | ||
| 11 | SERPINE2 | 1416666_at | 1993 | 1.733 | 0.0216 | No | ||
| 12 | TKTL1 | 1420064_s_at 1420065_at 1449567_at | 2055 | 1.695 | 0.0243 | No | ||
| 13 | H1F0 | 1423702_at 1450522_a_at | 2146 | 1.649 | 0.0256 | No | ||
| 14 | NPR3 | 1435184_at 1448024_at 1450286_at | 2204 | 1.617 | 0.0282 | No | ||
| 15 | MTMR11 | 1435294_at 1441835_x_at 1460550_at | 2338 | 1.548 | 0.0271 | No | ||
| 16 | TPSAB1 | 1426175_a_at | 2431 | 1.509 | 0.0278 | No | ||
| 17 | DLK1 | 1449939_s_at | 2579 | 1.442 | 0.0257 | No | ||
| 18 | BZRAP1 | 1455771_at | 2770 | 1.381 | 0.0215 | No | ||
| 19 | RAP2A | 1421574_at 1426965_at 1440473_at | 2993 | 1.314 | 0.0155 | No | ||
| 20 | TOX | 1425483_at 1425484_at 1442039_at 1446950_at 1458974_at | 3115 | 1.277 | 0.0141 | No | ||
| 21 | PALM | 1422034_a_at 1423967_at 1439927_at 1457217_at | 3134 | 1.272 | 0.0174 | No | ||
| 22 | TNFRSF21 | 1422740_at 1450731_s_at | 3218 | 1.251 | 0.0177 | No | ||
| 23 | MEF2A | 1421252_a_at 1425426_a_at 1427185_at 1427186_a_at 1452347_at | 3313 | 1.230 | 0.0174 | No | ||
| 24 | UGCG | 1421268_at 1421269_at 1435133_at 1439863_at | 3350 | 1.222 | 0.0197 | No | ||
| 25 | MPL | 1421461_at 1451919_a_at | 3418 | 1.208 | 0.0206 | No | ||
| 26 | ARHGEF17 | 1419938_s_at 1419939_at 1433682_at 1442357_at 1447441_at | 3749 | 1.140 | 0.0091 | No | ||
| 27 | PXDN | 1428259_at | 3922 | 1.111 | 0.0048 | No | ||
| 28 | SERPINF1 | 1416168_at 1453724_a_at | 4065 | 1.084 | 0.0018 | No | ||
| 29 | MS4A3 | 1420572_at | 4083 | 1.080 | 0.0045 | No | ||
| 30 | SPON1 | 1424415_s_at 1441226_at 1451342_at | 4273 | 1.050 | -0.0008 | No | ||
| 31 | GNAI1 | 1427510_at 1434440_at 1454959_s_at | 4341 | 1.041 | -0.0005 | No | ||
| 32 | LMO4 | 1420981_a_at 1439651_at | 4400 | 1.032 | 0.0002 | No | ||
| 33 | IGFBP7 | 1423584_at 1437804_at | 4469 | 1.024 | 0.0004 | No | ||
| 34 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 4473 | 1.023 | 0.0036 | No | ||
| 35 | DPYSL2 | 1433770_at 1450502_at 1458188_at 1458632_at | 4572 | 1.013 | 0.0024 | No | ||
| 36 | DCHS1 | 1429163_at | 4694 | 1.001 | 0.0001 | No | ||
| 37 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 4714 | 0.999 | 0.0025 | No | ||
| 38 | ARHGEF12 | 1423902_s_at 1446965_at 1451159_at 1453853_a_at | 4804 | 0.989 | 0.0016 | No | ||
| 39 | TFPI | 1438530_at 1451790_a_at 1451791_at 1452432_at | 4853 | 0.982 | 0.0026 | No | ||
| 40 | FLNB | 1424828_a_at 1426750_at 1442107_at 1445534_at 1458226_at | 4997 | 0.966 | -0.0008 | No | ||
| 41 | RUNX3 | 1421467_at 1440275_at | 5034 | 0.961 | 0.0006 | No | ||
| 42 | MMP9 | 1416298_at 1448291_at | 5171 | 0.941 | -0.0026 | No | ||
| 43 | GYPC | 1423878_at | 5176 | 0.941 | 0.0003 | No | ||
| 44 | HGF | 1425379_at 1442884_at 1451866_a_at | 5189 | 0.939 | 0.0028 | No | ||
| 45 | SCRN1 | 1428717_at 1428718_at | 5208 | 0.935 | 0.0050 | No | ||
| 46 | P2RX5 | 1449433_at | 5670 | 0.874 | -0.0133 | No | ||
| 47 | EMP1 | 1416529_at 1459171_at | 5909 | 0.842 | -0.0215 | No | ||
| 48 | CACNA2D2 | 1426185_at | 5921 | 0.841 | -0.0193 | No | ||
| 49 | PROM1 | 1419700_a_at | 6194 | 0.805 | -0.0292 | No | ||
| 50 | SYNGR1 | 1419289_a_at 1434661_at 1453772_at | 6282 | 0.795 | -0.0306 | No | ||
| 51 | CYP2E1 | 1415994_at | 6549 | 0.762 | -0.0404 | No | ||
| 52 | CIITA | 1421210_at 1421211_a_at 1447075_at | 6576 | 0.759 | -0.0391 | No | ||
| 53 | AGTPBP1 | 1429933_at 1432385_a_at | 7107 | 0.698 | -0.0612 | No | ||
| 54 | NBL1 | 1448428_at | 7122 | 0.696 | -0.0596 | No | ||
| 55 | BAIAP3 | 1427509_at | 7130 | 0.696 | -0.0577 | No | ||
| 56 | EGFL7 | 1421335_a_at 1435823_x_at 1451427_a_at 1451428_x_at | 7361 | 0.667 | -0.0661 | No | ||
| 57 | MYH11 | 1418122_at 1448962_at | 7383 | 0.665 | -0.0649 | No | ||
| 58 | APP | 1420621_a_at 1427442_a_at 1438373_at 1438374_x_at 1444705_at 1447347_at 1458525_at | 7461 | 0.655 | -0.0663 | No | ||
| 59 | CLEC5A | 1421366_at | 7473 | 0.654 | -0.0647 | No | ||
| 60 | RPS6KA2 | 1417542_at 1417543_at 1441311_at | 7640 | 0.637 | -0.0702 | No | ||
| 61 | SERPING1 | 1416625_at | 7933 | 0.607 | -0.0817 | No | ||
| 62 | AK3L1 | 1421829_at 1421830_at 1450387_s_at | 7997 | 0.600 | -0.0826 | No | ||
| 63 | FCER1A | 1421775_at | 8148 | 0.585 | -0.0876 | No | ||
| 64 | MST1 | 1418267_at | 8382 | 0.562 | -0.0965 | No | ||
| 65 | PTRF | 1421431_at 1424130_a_at 1453891_at | 8660 | 0.534 | -0.1075 | No | ||
| 66 | SNCA | 1418493_a_at 1431022_at 1436853_a_at 1446391_at | 9069 | 0.493 | -0.1247 | No | ||
| 67 | GPSM2 | 1424895_at | 9189 | 0.481 | -0.1286 | No | ||
| 68 | FHL1 | 1417872_at 1459003_at | 9442 | 0.457 | -0.1387 | No | ||
| 69 | RHOBTB3 | 1429661_at 1433647_s_at 1447869_x_at 1453227_at 1457009_at | 9448 | 0.457 | -0.1375 | No | ||
| 70 | MN1 | 1454867_at | 9590 | 0.441 | -0.1425 | No | ||
| 71 | RETN | 1449182_at | 9827 | 0.419 | -0.1520 | No | ||
| 72 | IL5RA | 1421620_at | 9850 | 0.417 | -0.1517 | No | ||
| 73 | GPA33 | 1419330_a_at 1454247_a_at | 9935 | 0.408 | -0.1542 | No | ||
| 74 | RUNX1T1 | 1427640_a_at 1437784_at 1440310_at 1443788_at 1444615_x_at 1444904_at 1448785_at | 10175 | 0.385 | -0.1639 | No | ||
| 75 | TCF3 | 1436207_at 1450117_at | 10239 | 0.379 | -0.1656 | No | ||
| 76 | P2RY5 | 1428615_at 1444627_at | 10367 | 0.368 | -0.1702 | No | ||
| 77 | TRH | 1418756_at | 10937 | 0.313 | -0.1954 | No | ||
| 78 | LRP3 | 1436083_at | 11044 | 0.304 | -0.1993 | No | ||
| 79 | MSLN | 1460238_at | 11056 | 0.303 | -0.1988 | No | ||
| 80 | TGFB1I1 | 1418136_at | 11076 | 0.302 | -0.1987 | No | ||
| 81 | ERG | 1425370_a_at 1440244_at | 11523 | 0.258 | -0.2184 | No | ||
| 82 | DACH1 | 1420694_a_at 1420695_at 1433743_at 1447174_at 1459381_at | 11669 | 0.242 | -0.2243 | No | ||
| 83 | BLNK | 1451780_at | 11797 | 0.229 | -0.2294 | No | ||
| 84 | CCR2 | 1421186_at 1421187_at 1421188_at 1460067_at | 11984 | 0.210 | -0.2372 | No | ||
| 85 | TMOD1 | 1419863_at 1422754_at | 11998 | 0.208 | -0.2372 | No | ||
| 86 | FGFR1 | 1424050_s_at 1425911_a_at 1436551_at | 12054 | 0.204 | -0.2390 | No | ||
| 87 | ZAP70 | 1422701_at 1439749_at 1440178_x_at | 12793 | 0.138 | -0.2725 | No | ||
| 88 | EVI1 | 1423011_at 1438325_at | 12843 | 0.132 | -0.2744 | No | ||
| 89 | LHFPL2 | 1434129_s_at 1434130_at 1443185_at 1454847_at | 12866 | 0.129 | -0.2749 | No | ||
| 90 | ST18 | 1436793_at 1455123_at | 12940 | 0.122 | -0.2779 | No | ||
| 91 | MEST | 1423294_at | 13253 | 0.087 | -0.2920 | No | ||
| 92 | VLDLR | 1417900_a_at 1434465_x_at 1435893_at 1438258_at 1442169_at 1451156_s_at | 13272 | 0.085 | -0.2925 | No | ||
| 93 | B4GALT6 | 1441169_at | 13282 | 0.083 | -0.2927 | No | ||
| 94 | CAV1 | 1449145_a_at | 13411 | 0.070 | -0.2983 | No | ||
| 95 | ALDH2 | 1434987_at 1434988_x_at 1437410_at 1448143_at | 13489 | 0.061 | -0.3017 | No | ||
| 96 | EPS8 | 1422823_at 1422824_s_at 1425733_a_at 1436977_at | 13587 | 0.050 | -0.3060 | No | ||
| 97 | SART2 | 1455795_at | 13673 | 0.042 | -0.3098 | No | ||
| 98 | ROBO1 | 1427231_at 1443042_at 1457407_at 1459453_at | 13676 | 0.042 | -0.3097 | No | ||
| 99 | POU4F1 | 1429667_at 1429668_at | 13756 | 0.033 | -0.3132 | No | ||
| 100 | NARFL | 1450452_a_at | 13789 | 0.029 | -0.3146 | No | ||
| 101 | BAALC | 1426329_s_at 1430649_at 1436878_at 1452022_at | 13807 | 0.027 | -0.3153 | No | ||
| 102 | NRXN2 | 1435907_at 1435908_at 1455499_at | 14039 | 0.003 | -0.3259 | No | ||
| 103 | F2RL1 | 1448931_at | 14169 | -0.013 | -0.3318 | No | ||
| 104 | WASF1 | 1418545_at 1440438_at | 14215 | -0.017 | -0.3338 | No | ||
| 105 | CLDN15 | 1418920_at | 14246 | -0.021 | -0.3352 | No | ||
| 106 | SPRY1 | 1415874_at | 14537 | -0.058 | -0.3483 | No | ||
| 107 | ITM2C | 1415961_at | 14547 | -0.059 | -0.3485 | No | ||
| 108 | FZD6 | 1417301_at 1448662_at 1458904_at | 14607 | -0.066 | -0.3510 | No | ||
| 109 | NT5E | 1416517_at 1422974_at 1427379_at 1428547_at | 14639 | -0.070 | -0.3522 | No | ||
| 110 | CD19 | 1450570_a_at | 14726 | -0.083 | -0.3559 | No | ||
| 111 | HPGD | 1419905_s_at 1419906_at | 14784 | -0.089 | -0.3582 | No | ||
| 112 | RGS1 | 1417601_at | 14869 | -0.100 | -0.3618 | No | ||
| 113 | CD34 | 1416072_at | 15048 | -0.124 | -0.3696 | No | ||
| 114 | SCCPDH | 1426510_at 1438909_at | 15177 | -0.140 | -0.3750 | No | ||
| 115 | TRPM4 | 1435549_at | 15304 | -0.159 | -0.3803 | No | ||
| 116 | PTPRM | 1422541_at 1441199_at | 15720 | -0.221 | -0.3987 | No | ||
| 117 | FAIM3 | 1429889_at | 15831 | -0.239 | -0.4029 | No | ||
| 118 | RBPMS | 1425652_s_at 1429359_s_at 1437161_x_at 1444765_at 1455936_a_at 1459078_at | 15844 | -0.241 | -0.4027 | No | ||
| 119 | SLC38A1 | 1415903_at 1454764_s_at | 15849 | -0.242 | -0.4021 | No | ||
| 120 | FZD2 | 1418532_at 1418533_s_at | 15934 | -0.261 | -0.4051 | No | ||
| 121 | SNRPN /// SNURF | 1421063_s_at | 15958 | -0.266 | -0.4053 | No | ||
| 122 | NCAM1 | 1421966_at 1425126_at 1426864_a_at 1426865_a_at 1439556_at 1440067_at 1441995_at 1442680_at 1443018_at 1450437_a_at 1450438_at 1454140_at | 16044 | -0.280 | -0.4083 | No | ||
| 123 | BANK1 | 1442646_at 1456328_at | 16204 | -0.310 | -0.4146 | No | ||
| 124 | ICAM4 | 1422930_at | 16220 | -0.313 | -0.4143 | No | ||
| 125 | GATM | 1423569_at | 16274 | -0.325 | -0.4157 | No | ||
| 126 | DEPDC6 | 1419963_at 1428622_at 1443579_s_at 1446254_at 1451348_at 1453571_at 1459719_at | 16409 | -0.350 | -0.4207 | No | ||
| 127 | ADCY3 | 1421959_s_at 1421960_at 1436928_s_at 1436929_x_at 1440405_at 1444641_at | 16444 | -0.358 | -0.4211 | No | ||
| 128 | STK32B | 1431236_at 1452420_at | 16505 | -0.370 | -0.4227 | No | ||
| 129 | YPEL1 | 1421048_a_at 1431781_at 1431782_s_at 1442660_at | 16571 | -0.385 | -0.4244 | No | ||
| 130 | CD38 | 1433741_at 1450136_at | 16624 | -0.398 | -0.4255 | No | ||
| 131 | CSF3R | 1418806_at | 16734 | -0.430 | -0.4291 | No | ||
| 132 | KCNE1L | 1416640_at | 16882 | -0.466 | -0.4344 | No | ||
| 133 | OPTN | 1429778_at 1435679_at | 16943 | -0.481 | -0.4356 | No | ||
| 134 | IFITM1 | 1424254_at | 17040 | -0.504 | -0.4383 | No | ||
| 135 | TACSTD2 | 1423323_at | 17111 | -0.524 | -0.4398 | No | ||
| 136 | SH3BP4 | 1416305_at 1455250_at | 17135 | -0.529 | -0.4392 | No | ||
| 137 | GJA1 | 1415800_at 1415801_at 1437992_x_at 1438650_x_at 1438945_x_at 1438973_x_at | 17261 | -0.570 | -0.4431 | No | ||
| 138 | CENTD1 | 1436895_at 1446649_at 1452291_at 1456337_at 1458173_at | 17442 | -0.621 | -0.4493 | No | ||
| 139 | FOXO1A | 1416981_at 1416982_at 1416983_s_at 1459170_at | 17552 | -0.660 | -0.4522 | No | ||
| 140 | C1ORF24 | 1422567_at 1454942_at | 17582 | -0.669 | -0.4514 | No | ||
| 141 | PTGIR | 1427313_at | 17737 | -0.710 | -0.4561 | No | ||
| 142 | DSU | 1437250_at | 17818 | -0.742 | -0.4574 | No | ||
| 143 | ITGA6 | 1422444_at 1422445_at | 18008 | -0.808 | -0.4635 | No | ||
| 144 | PMAIP1 | 1418203_at | 18072 | -0.835 | -0.4636 | No | ||
| 145 | ENDOD1 | 1426540_at 1426541_a_at 1426542_at 1426543_x_at 1433796_at | 18148 | -0.864 | -0.4643 | No | ||
| 146 | IFI44 | 1423555_a_at | 18215 | -0.886 | -0.4644 | No | ||
| 147 | HSPB1 | 1422943_a_at 1425964_x_at 1427853_a_at 1427854_x_at 1453914_at | 18292 | -0.911 | -0.4650 | No | ||
| 148 | PRKD3 | 1420567_at 1428229_at 1428230_at 1444691_at | 18552 | -0.994 | -0.4736 | No | ||
| 149 | RRAGD | 1431164_at 1434909_at 1440555_at | 18707 | -1.040 | -0.4773 | No | ||
| 150 | CHI3L1 | 1451537_at | 18875 | -1.110 | -0.4814 | Yes | ||
| 151 | BPGM | 1415864_at 1415865_s_at 1448119_at | 18897 | -1.119 | -0.4787 | Yes | ||
| 152 | ELA2 | 1422928_at | 18910 | -1.124 | -0.4756 | Yes | ||
| 153 | FGF13 | 1418497_at 1418498_at | 19002 | -1.169 | -0.4760 | Yes | ||
| 154 | GPM6B | 1423091_a_at 1425942_a_at 1453514_at | 19201 | -1.263 | -0.4810 | Yes | ||
| 155 | MLLT3 | 1429205_at 1431890_a_at 1441535_at 1453622_s_at | 19252 | -1.293 | -0.4791 | Yes | ||
| 156 | VIL2 | 1450850_at 1456725_x_at | 19287 | -1.312 | -0.4764 | Yes | ||
| 157 | CD48 | 1427301_at | 19435 | -1.398 | -0.4786 | Yes | ||
| 158 | PAM | 1418908_at 1445797_at | 19450 | -1.409 | -0.4747 | Yes | ||
| 159 | PTPRCAP | 1448511_at | 19621 | -1.514 | -0.4776 | Yes | ||
| 160 | VEGF | 1420909_at 1451959_a_at | 19669 | -1.555 | -0.4747 | Yes | ||
| 161 | TM6SF1 | 1451353_at | 19938 | -1.754 | -0.4813 | Yes | ||
| 162 | LAMC1 | 1423885_at 1423886_at | 20064 | -1.855 | -0.4810 | Yes | ||
| 163 | RAMP1 | 1417481_at 1447474_at | 20122 | -1.899 | -0.4774 | Yes | ||
| 164 | PPP1R16B | 1425414_at 1455080_at | 20187 | -1.959 | -0.4740 | Yes | ||
| 165 | VPREB1 | 1449869_at | 20202 | -1.969 | -0.4682 | Yes | ||
| 166 | GSN | 1415812_at 1436991_x_at 1437171_x_at 1441225_at 1456312_x_at 1456568_at 1456569_x_at | 20219 | -1.981 | -0.4625 | Yes | ||
| 167 | SPARC | 1416589_at 1448392_at 1458204_at | 20272 | -2.010 | -0.4584 | Yes | ||
| 168 | PTGER4 | 1421073_a_at 1424208_at 1436441_at | 20389 | -2.086 | -0.4569 | Yes | ||
| 169 | CD200 | 1448788_at 1458082_at | 20641 | -2.382 | -0.4607 | Yes | ||
| 170 | GMPR | 1448530_at | 20643 | -2.383 | -0.4530 | Yes | ||
| 171 | TSPAN13 | 1418643_at 1460239_at | 20790 | -2.562 | -0.4514 | Yes | ||
| 172 | MDFIC | 1427040_at | 20842 | -2.645 | -0.4451 | Yes | ||
| 173 | DNTT | 1420166_at 1449757_x_at 1450545_a_at | 20863 | -2.683 | -0.4373 | Yes | ||
| 174 | TGFBI | 1415871_at 1437463_x_at 1448123_s_at 1456250_x_at | 20889 | -2.720 | -0.4296 | Yes | ||
| 175 | PPP4R2 | 1433851_at 1442834_at | 20949 | -2.801 | -0.4232 | Yes | ||
| 176 | PDE3B | 1433694_at | 20971 | -2.853 | -0.4149 | Yes | ||
| 177 | SERINC5 | 1433571_at | 21101 | -3.129 | -0.4106 | Yes | ||
| 178 | SORL1 | 1426258_at 1445889_at 1453003_at 1460390_at | 21122 | -3.162 | -0.4013 | Yes | ||
| 179 | SIDT1 | 1426550_at 1426551_at | 21258 | -3.519 | -0.3960 | Yes | ||
| 180 | PRTN3 | 1419669_at | 21264 | -3.531 | -0.3848 | Yes | ||
| 181 | ZHX2 | 1455210_at | 21272 | -3.557 | -0.3735 | Yes | ||
| 182 | MPO | 1415960_at | 21304 | -3.651 | -0.3630 | Yes | ||
| 183 | GATA3 | 1448886_at | 21331 | -3.739 | -0.3521 | Yes | ||
| 184 | SPIB | 1460407_at | 21336 | -3.766 | -0.3400 | Yes | ||
| 185 | ALAS2 | 1451675_a_at | 21415 | -4.037 | -0.3304 | Yes | ||
| 186 | DHRS3 | 1448390_a_at | 21429 | -4.091 | -0.3177 | Yes | ||
| 187 | SSBP2 | 1429951_at 1438423_at 1449815_a_at 1451542_at | 21486 | -4.320 | -0.3062 | Yes | ||
| 188 | NEDD4 | 1421955_a_at 1444044_at 1450431_a_at 1451109_a_at | 21487 | -4.321 | -0.2922 | Yes | ||
| 189 | CD3D | 1422828_at | 21560 | -4.763 | -0.2800 | Yes | ||
| 190 | CD69 | 1428735_at | 21570 | -4.845 | -0.2646 | Yes | ||
| 191 | JUP | 1426873_s_at | 21633 | -5.336 | -0.2501 | Yes | ||
| 192 | CD7 | 1419711_at | 21644 | -5.423 | -0.2329 | Yes | ||
| 193 | LCN2 | 1427747_a_at | 21666 | -5.583 | -0.2157 | Yes | ||
| 194 | ABLIM1 | 1442376_at 1453103_at 1454708_at 1460120_at | 21696 | -5.811 | -0.1981 | Yes | ||
| 195 | CD2 | 1418770_at | 21715 | -6.027 | -0.1793 | Yes | ||
| 196 | EHD3 | 1417235_at 1417236_at | 21721 | -6.095 | -0.1597 | Yes | ||
| 197 | ISG20 | 1419569_a_at | 21753 | -6.425 | -0.1402 | Yes | ||
| 198 | PLXND1 | 1451475_at | 21814 | -7.199 | -0.1196 | Yes | ||
| 199 | ITM2A | 1423608_at 1451047_at | 21840 | -7.488 | -0.0963 | Yes | ||
| 200 | ADIPOR1 | 1424312_at 1439017_x_at 1451311_a_at | 21855 | -7.652 | -0.0721 | Yes | ||
| 201 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21869 | -7.737 | -0.0475 | Yes | ||
| 202 | PDE4B | 1422473_at 1422474_at 1442700_at 1444731_at 1445449_at 1446577_at 1447237_at 1447718_at | 21873 | -7.745 | -0.0224 | Yes | ||
| 203 | EVL | 1434920_a_at 1440885_at 1445957_at 1450106_a_at 1456453_at 1458889_at | 21877 | -7.764 | 0.0027 | Yes |