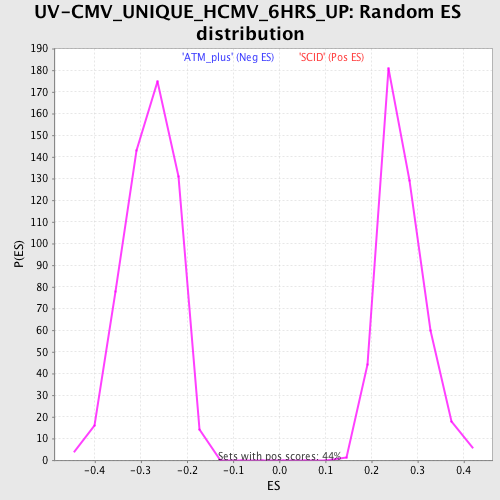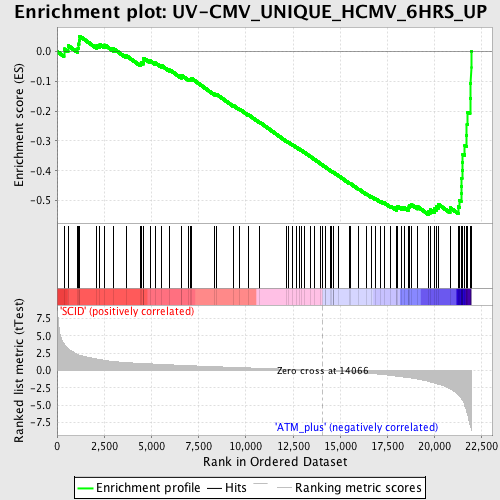
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | UV-CMV_UNIQUE_HCMV_6HRS_UP |
| Enrichment Score (ES) | -0.5464862 |
| Normalized Enrichment Score (NES) | -1.9568374 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0118586365 |
| FWER p-Value | 0.137 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHEK2 | 1422747_at | 365 | 3.836 | 0.0085 | No | ||
| 2 | RQCD1 | 1450887_at | 578 | 3.231 | 0.0201 | No | ||
| 3 | UBE2S | 1416726_s_at 1430962_at | 1103 | 2.316 | 0.0113 | No | ||
| 4 | MX2 | 1419676_at | 1129 | 2.284 | 0.0252 | No | ||
| 5 | TRADD | 1429117_at 1439910_a_at 1452622_a_at | 1160 | 2.241 | 0.0386 | No | ||
| 6 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 1195 | 2.213 | 0.0516 | No | ||
| 7 | NP | 1416530_a_at | 2106 | 1.668 | 0.0209 | No | ||
| 8 | ACTL6B | 1422564_at | 2269 | 1.583 | 0.0239 | No | ||
| 9 | AREG | 1421134_at | 2526 | 1.463 | 0.0218 | No | ||
| 10 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 2977 | 1.318 | 0.0099 | No | ||
| 11 | BRCA2 | 1419076_a_at | 3648 | 1.156 | -0.0132 | No | ||
| 12 | CH25H | 1449227_at | 4422 | 1.030 | -0.0418 | No | ||
| 13 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 4473 | 1.023 | -0.0374 | No | ||
| 14 | TLR3 | 1422781_at 1422782_s_at | 4575 | 1.012 | -0.0353 | No | ||
| 15 | TREX1 | 1450672_a_at 1459618_at | 4585 | 1.011 | -0.0291 | No | ||
| 16 | STX11 | 1423576_a_at 1451032_at 1453228_at 1457780_at | 4587 | 1.011 | -0.0225 | No | ||
| 17 | HBEGF | 1418349_at 1418350_at | 4924 | 0.976 | -0.0315 | No | ||
| 18 | DPT | 1418511_at | 5207 | 0.936 | -0.0382 | No | ||
| 19 | CRYGD | 1419141_at | 5528 | 0.892 | -0.0470 | No | ||
| 20 | IL1RAP | 1421843_at 1439697_at 1442614_at 1449585_at | 5971 | 0.835 | -0.0617 | No | ||
| 21 | RGS6 | 1427340_at 1452399_at | 6571 | 0.759 | -0.0842 | No | ||
| 22 | CCL8 | 1419684_at | 6602 | 0.756 | -0.0805 | No | ||
| 23 | TNFAIP3 | 1433699_at 1450829_at | 6966 | 0.715 | -0.0925 | No | ||
| 24 | ADAMTS3 | 1441693_at 1458143_at | 7080 | 0.701 | -0.0930 | No | ||
| 25 | AGTPBP1 | 1429933_at 1432385_a_at | 7107 | 0.698 | -0.0896 | No | ||
| 26 | INDO | 1420437_at | 8341 | 0.567 | -0.1423 | No | ||
| 27 | PDE1A | 1431625_at 1449298_a_at | 8453 | 0.555 | -0.1438 | No | ||
| 28 | EPHA5 | 1420557_at 1437920_at 1444690_at | 9318 | 0.469 | -0.1802 | No | ||
| 29 | OAS2 | 1425065_at | 9664 | 0.434 | -0.1932 | No | ||
| 30 | RABGAP1L | 1421488_at 1429196_at 1429197_s_at 1434062_at 1441202_at 1453365_at 1457178_at 1458155_at | 10130 | 0.390 | -0.2119 | No | ||
| 31 | IGFBP2 | 1454159_a_at | 10729 | 0.332 | -0.2371 | No | ||
| 32 | ICAM1 | 1424067_at | 12150 | 0.196 | -0.3008 | No | ||
| 33 | ELN | 1420854_at 1420855_at 1446221_at | 12259 | 0.186 | -0.3045 | No | ||
| 34 | TROVE2 | 1423433_at 1436533_at 1436534_at 1436535_at | 12462 | 0.167 | -0.3127 | No | ||
| 35 | TAC1 | 1416783_at 1419411_at 1431883_at | 12659 | 0.150 | -0.3206 | No | ||
| 36 | PLA2G4A | 1448558_a_at | 12813 | 0.137 | -0.3267 | No | ||
| 37 | PCDH9 | 1439322_at 1442659_at 1444215_at 1444724_at 1445052_at 1445061_at 1457973_at | 12927 | 0.124 | -0.3311 | No | ||
| 38 | EGFR | 1424932_at 1432647_at 1435888_at 1451530_at 1454313_at 1457563_at 1460420_a_at | 13105 | 0.105 | -0.3385 | No | ||
| 39 | CPEB3 | 1437765_at 1445868_at 1455372_at 1456048_at | 13415 | 0.069 | -0.3522 | No | ||
| 40 | COL9A3 | 1420280_x_at 1460693_a_at 1460734_at | 13617 | 0.047 | -0.3611 | No | ||
| 41 | GALNT3 | 1417588_at | 13948 | 0.013 | -0.3761 | No | ||
| 42 | CCL5 | 1418126_at | 14078 | -0.001 | -0.3820 | No | ||
| 43 | NFKB2 | 1425902_a_at 1429128_x_at | 14195 | -0.015 | -0.3872 | No | ||
| 44 | MYD88 | 1419272_at | 14240 | -0.021 | -0.3891 | No | ||
| 45 | FGFR3 | 1421841_at 1425796_a_at | 14464 | -0.049 | -0.3990 | No | ||
| 46 | KIF5C | 1422945_a_at 1427635_at 1447454_at 1450804_at 1455266_at 1457255_x_at | 14513 | -0.055 | -0.4008 | No | ||
| 47 | TNFSF10 | 1420412_at 1439680_at 1459913_at | 14624 | -0.069 | -0.4054 | No | ||
| 48 | TDRD7 | 1426716_at 1459216_at | 14630 | -0.070 | -0.4052 | No | ||
| 49 | IL6 | 1450297_at | 14660 | -0.073 | -0.4060 | No | ||
| 50 | TFEC | 1419537_at | 14906 | -0.104 | -0.4166 | No | ||
| 51 | FGF2 | 1449826_a_at | 15498 | -0.189 | -0.4424 | No | ||
| 52 | AUTS2 | 1427293_a_at 1438680_at 1441869_x_at 1442360_at 1443062_at 1443070_at 1443770_x_at 1444069_at 1445307_at 1445469_at 1445538_at 1446849_at 1452379_at 1457139_at 1458592_at 1459338_at | 15520 | -0.193 | -0.4421 | No | ||
| 53 | COL15A1 | 1448755_at | 15988 | -0.270 | -0.4617 | No | ||
| 54 | CCL19 | 1449277_at | 16390 | -0.347 | -0.4777 | No | ||
| 55 | CD38 | 1433741_at 1450136_at | 16624 | -0.398 | -0.4858 | No | ||
| 56 | CX3CR1 | 1450019_at 1450020_at | 16876 | -0.464 | -0.4942 | No | ||
| 57 | ING1 | 1416860_s_at 1433267_at 1448496_a_at 1454534_at | 17142 | -0.530 | -0.5029 | No | ||
| 58 | MSX1 | 1417127_at 1448601_s_at | 17320 | -0.585 | -0.5071 | No | ||
| 59 | GEM | 1426063_a_at | 17669 | -0.693 | -0.5185 | No | ||
| 60 | DOPEY2 | 1428330_at 1449904_at | 17978 | -0.795 | -0.5274 | No | ||
| 61 | BAZ1A | 1433599_at 1447930_at | 17979 | -0.796 | -0.5221 | No | ||
| 62 | RBBP6 | 1425114_at 1425115_at 1425421_at 1426487_a_at 1453611_at | 18045 | -0.825 | -0.5197 | No | ||
| 63 | SOS1 | 1421884_at 1421885_at 1421886_at | 18257 | -0.899 | -0.5234 | No | ||
| 64 | SAMHD1 | 1418131_at 1434438_at 1444064_at | 18372 | -0.935 | -0.5225 | No | ||
| 65 | PFKFB2 | 1422090_a_at 1422091_at 1422092_at 1429486_at 1431901_a_at | 18596 | -1.004 | -0.5261 | No | ||
| 66 | CREM | 1418322_at 1430598_at 1430847_a_at 1449037_at | 18635 | -1.019 | -0.5211 | No | ||
| 67 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 18671 | -1.030 | -0.5160 | No | ||
| 68 | BAMBI | 1423753_at | 18760 | -1.063 | -0.5130 | No | ||
| 69 | GTF2B | 1438066_at 1451135_at | 19091 | -1.206 | -0.5202 | No | ||
| 70 | TNFSF9 | 1422924_at | 19667 | -1.554 | -0.5363 | Yes | ||
| 71 | TAF13 | 1448707_at | 19786 | -1.637 | -0.5309 | Yes | ||
| 72 | IRF4 | 1421173_at 1421174_at | 19973 | -1.781 | -0.5277 | Yes | ||
| 73 | TRIM21 | 1418077_at 1448940_at | 20080 | -1.868 | -0.5203 | Yes | ||
| 74 | PPP1R16B | 1425414_at 1455080_at | 20187 | -1.959 | -0.5122 | Yes | ||
| 75 | ELF1 | 1417540_at 1439319_at | 20812 | -2.600 | -0.5237 | Yes | ||
| 76 | IRF7 | 1417244_a_at | 21245 | -3.481 | -0.5206 | Yes | ||
| 77 | IER2 | 1416442_at | 21321 | -3.698 | -0.4997 | Yes | ||
| 78 | SOCS1 | 1440047_at 1450446_a_at | 21397 | -3.962 | -0.4771 | Yes | ||
| 79 | CCRK | 1422494_s_at | 21406 | -4.013 | -0.4510 | Yes | ||
| 80 | DNAJB9 | 1417191_at | 21432 | -4.103 | -0.4252 | Yes | ||
| 81 | DUSP2 | 1450698_at | 21446 | -4.159 | -0.3984 | Yes | ||
| 82 | JUNB | 1415899_at | 21479 | -4.278 | -0.3718 | Yes | ||
| 83 | RRAD | 1422562_at | 21490 | -4.331 | -0.3437 | Yes | ||
| 84 | RELB | 1417856_at | 21596 | -5.083 | -0.3151 | Yes | ||
| 85 | CSTF2 | 1419644_at 1419645_at 1455523_at | 21682 | -5.705 | -0.2815 | Yes | ||
| 86 | CRY1 | 1433733_a_at | 21708 | -5.979 | -0.2433 | Yes | ||
| 87 | ISG20 | 1419569_a_at | 21753 | -6.425 | -0.2030 | Yes | ||
| 88 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21869 | -7.737 | -0.1574 | Yes | ||
| 89 | IFI27 | 1426278_at | 21901 | -7.917 | -0.1068 | Yes | ||
| 90 | EGR2 | 1427682_a_at 1427683_at | 21921 | -8.208 | -0.0536 | Yes | ||
| 91 | EGR3 | 1421486_at 1436329_at | 21923 | -8.252 | 0.0006 | Yes |