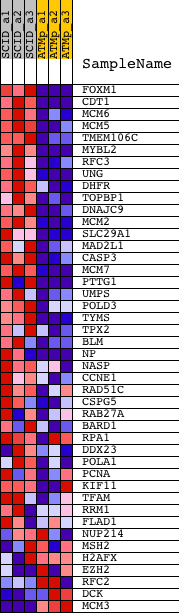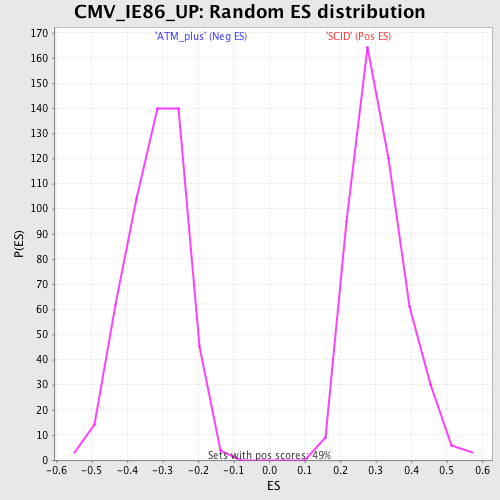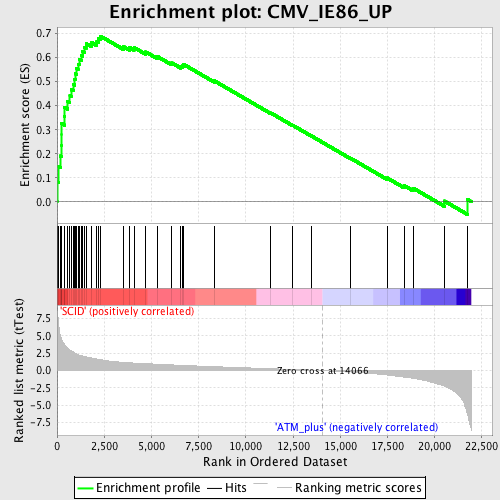
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | CMV_IE86_UP |
| Enrichment Score (ES) | 0.6899395 |
| Normalized Enrichment Score (NES) | 2.2370057 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FOXM1 | 1417748_x_at 1437138_at 1448833_at 1448834_at | 4 | 8.471 | 0.0838 | Yes | ||
| 2 | CDT1 | 1424143_a_at 1424144_at 1455791_at | 87 | 6.599 | 0.1454 | Yes | ||
| 3 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 182 | 5.000 | 0.1907 | Yes | ||
| 4 | MCM5 | 1415945_at 1436808_x_at | 216 | 4.655 | 0.2353 | Yes | ||
| 5 | TMEM106C | 1434783_at 1434784_s_at | 223 | 4.604 | 0.2807 | Yes | ||
| 6 | MYBL2 | 1417656_at 1454946_at | 225 | 4.592 | 0.3261 | Yes | ||
| 7 | RFC3 | 1423700_at 1432538_a_at | 381 | 3.777 | 0.3565 | Yes | ||
| 8 | UNG | 1425753_a_at | 393 | 3.740 | 0.3931 | Yes | ||
| 9 | DHFR | 1419172_at 1430750_at | 556 | 3.278 | 0.4182 | Yes | ||
| 10 | TOPBP1 | 1452241_at 1458480_at | 680 | 2.980 | 0.4421 | Yes | ||
| 11 | DNAJC9 | 1426473_at | 744 | 2.841 | 0.4673 | Yes | ||
| 12 | MCM2 | 1434079_s_at 1448777_at | 864 | 2.652 | 0.4882 | Yes | ||
| 13 | SLC29A1 | 1451782_a_at | 937 | 2.532 | 0.5100 | Yes | ||
| 14 | MAD2L1 | 1422460_at | 959 | 2.501 | 0.5338 | Yes | ||
| 15 | CASP3 | 1426165_a_at 1430192_at 1449839_at | 1020 | 2.418 | 0.5550 | Yes | ||
| 16 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 1113 | 2.307 | 0.5737 | Yes | ||
| 17 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 1195 | 2.213 | 0.5919 | Yes | ||
| 18 | UMPS | 1431652_at 1434859_at 1451445_at 1455832_a_at | 1299 | 2.131 | 0.6084 | Yes | ||
| 19 | POLD3 | 1426838_at 1426839_at 1443733_x_at 1459647_at | 1340 | 2.099 | 0.6273 | Yes | ||
| 20 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 1457 | 2.019 | 0.6420 | Yes | ||
| 21 | TPX2 | 1428104_at 1428105_at 1447518_at 1447519_x_at | 1536 | 1.985 | 0.6582 | Yes | ||
| 22 | BLM | 1448953_at | 1833 | 1.821 | 0.6627 | Yes | ||
| 23 | NP | 1416530_a_at | 2106 | 1.668 | 0.6668 | Yes | ||
| 24 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 2179 | 1.627 | 0.6796 | Yes | ||
| 25 | CCNE1 | 1416492_at 1441910_x_at | 2295 | 1.570 | 0.6899 | Yes | ||
| 26 | RAD51C | 1423026_at 1438453_at | 3501 | 1.187 | 0.6467 | No | ||
| 27 | CSPG5 | 1450713_at | 3836 | 1.123 | 0.6425 | No | ||
| 28 | RAB27A | 1425284_a_at 1425285_a_at 1429123_at 1454438_at | 4074 | 1.082 | 0.6424 | No | ||
| 29 | BARD1 | 1420594_at | 4697 | 1.001 | 0.6239 | No | ||
| 30 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 5320 | 0.921 | 0.6047 | No | ||
| 31 | DDX23 | 1430050_at | 6080 | 0.822 | 0.5781 | No | ||
| 32 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 6551 | 0.762 | 0.5642 | No | ||
| 33 | PCNA | 1417947_at | 6659 | 0.750 | 0.5668 | No | ||
| 34 | KIF11 | 1435306_a_at 1452314_at 1452315_at | 6705 | 0.744 | 0.5721 | No | ||
| 35 | TFAM | 1448224_at 1456215_at | 8327 | 0.567 | 0.5037 | No | ||
| 36 | RRM1 | 1415878_at 1440073_at 1448127_at | 11318 | 0.279 | 0.3699 | No | ||
| 37 | FLAD1 | 1424421_at | 12464 | 0.167 | 0.3192 | No | ||
| 38 | NUP214 | 1434351_at 1456503_at 1457815_at | 13468 | 0.063 | 0.2740 | No | ||
| 39 | MSH2 | 1416988_at 1447888_x_at | 15547 | -0.196 | 0.1810 | No | ||
| 40 | H2AFX | 1416746_at | 17477 | -0.632 | 0.0992 | No | ||
| 41 | EZH2 | 1416544_at 1444263_at | 18393 | -0.943 | 0.0667 | No | ||
| 42 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 18857 | -1.102 | 0.0565 | No | ||
| 43 | DCK | 1428838_a_at 1439012_a_at 1449176_a_at | 20506 | -2.207 | 0.0031 | No | ||
| 44 | MCM3 | 1420029_at 1426653_at | 21738 | -6.271 | 0.0090 | No |