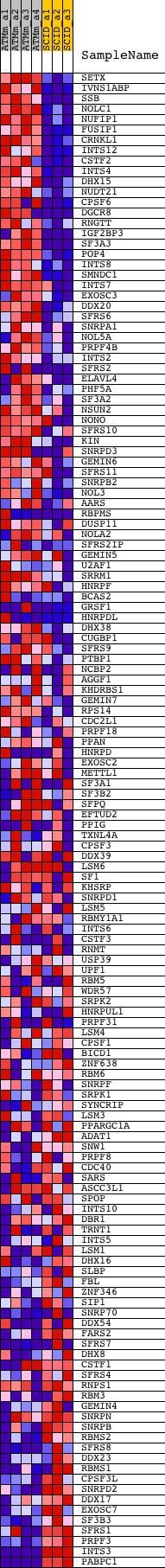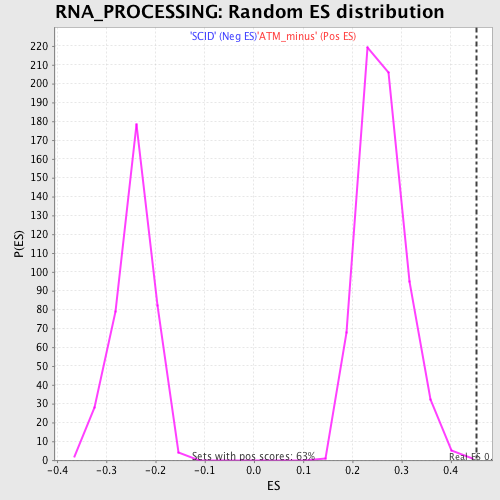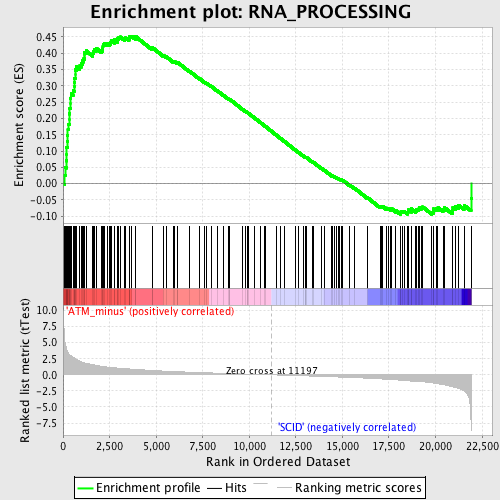
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | RNA_PROCESSING |
| Enrichment Score (ES) | 0.4524232 |
| Normalized Enrichment Score (NES) | 1.7354033 |
| Nominal p-value | 0.0015948963 |
| FDR q-value | 0.49224886 |
| FWER p-Value | 0.954 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SETX | 1433794_at 1440459_at 1458080_at | 89 | 5.411 | 0.0250 | Yes | ||
| 2 | IVNS1ABP | 1420961_a_at 1425718_a_at 1450084_s_at | 111 | 4.887 | 0.0503 | Yes | ||
| 3 | SSB | 1416421_a_at 1416422_a_at 1416423_x_at 1453928_a_at | 165 | 4.214 | 0.0705 | Yes | ||
| 4 | NOLC1 | 1428869_at 1428870_at 1450087_a_at | 190 | 3.938 | 0.0906 | Yes | ||
| 5 | NUFIP1 | 1448802_at | 196 | 3.885 | 0.1112 | Yes | ||
| 6 | FUSIP1 | 1418526_at 1418527_a_at 1423982_at 1449121_at | 229 | 3.646 | 0.1293 | Yes | ||
| 7 | CRNKL1 | 1420849_at 1420850_at | 241 | 3.591 | 0.1481 | Yes | ||
| 8 | INTS12 | 1428985_at 1452316_at | 261 | 3.474 | 0.1659 | Yes | ||
| 9 | CSTF2 | 1419644_at 1419645_at 1455523_at | 289 | 3.323 | 0.1826 | Yes | ||
| 10 | INTS4 | 1423806_at | 331 | 3.176 | 0.1977 | Yes | ||
| 11 | DHX15 | 1416144_a_at 1416145_at | 335 | 3.164 | 0.2146 | Yes | ||
| 12 | NUDT21 | 1417681_at 1428339_at 1437213_at 1455966_s_at | 361 | 3.083 | 0.2300 | Yes | ||
| 13 | CPSF6 | 1428231_at 1428232_at 1428233_at 1428234_at 1437372_at 1439188_at 1447381_at 1459687_x_at | 383 | 3.049 | 0.2455 | Yes | ||
| 14 | DGCR8 | 1422981_at 1435439_at 1446169_at 1455311_at | 386 | 3.047 | 0.2617 | Yes | ||
| 15 | RNGTT | 1425844_a_at 1448714_at | 423 | 2.974 | 0.2761 | Yes | ||
| 16 | IGF2BP3 | 1422610_s_at 1422611_s_at 1433731_at 1433732_x_at 1436145_at 1450687_at 1453957_a_at 1459016_at | 536 | 2.718 | 0.2855 | Yes | ||
| 17 | SF3A3 | 1423811_at | 594 | 2.610 | 0.2969 | Yes | ||
| 18 | POP4 | 1448419_at | 601 | 2.590 | 0.3106 | Yes | ||
| 19 | INTS8 | 1431096_at 1435250_at 1456775_at 1460609_at | 612 | 2.560 | 0.3239 | Yes | ||
| 20 | SMNDC1 | 1429043_at 1452999_at | 647 | 2.493 | 0.3357 | Yes | ||
| 21 | INTS7 | 1428531_at 1428532_at 1443045_at 1452822_at 1458590_at | 649 | 2.486 | 0.3490 | Yes | ||
| 22 | EXOSC3 | 1431502_a_at | 708 | 2.375 | 0.3591 | Yes | ||
| 23 | DDX20 | 1416751_a_at | 903 | 2.094 | 0.3615 | Yes | ||
| 24 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 990 | 1.984 | 0.3682 | Yes | ||
| 25 | SNRPA1 | 1417351_a_at 1417352_s_at 1417353_x_at | 1037 | 1.942 | 0.3765 | Yes | ||
| 26 | NOL5A | 1426533_at 1455035_s_at | 1117 | 1.866 | 0.3829 | Yes | ||
| 27 | PRPF4B | 1425497_a_at 1425498_at 1436427_at 1451732_at 1451909_a_at 1455696_a_at | 1144 | 1.843 | 0.3917 | Yes | ||
| 28 | INTS2 | 1429461_at 1436247_at | 1146 | 1.843 | 0.4015 | Yes | ||
| 29 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 1230 | 1.777 | 0.4073 | Yes | ||
| 30 | ELAVL4 | 1428741_at 1450258_a_at 1452894_at 1457399_at | 1599 | 1.560 | 0.3988 | Yes | ||
| 31 | PHF5A | 1424170_at | 1625 | 1.545 | 0.4059 | Yes | ||
| 32 | SF3A2 | 1450576_a_at 1455546_s_at | 1672 | 1.526 | 0.4120 | Yes | ||
| 33 | NSUN2 | 1423850_at 1453759_at | 1791 | 1.459 | 0.4144 | Yes | ||
| 34 | NONO | 1415820_x_at 1431239_at 1448103_s_at | 2049 | 1.332 | 0.4098 | Yes | ||
| 35 | SFRS10 | 1419543_a_at 1427432_a_at 1441466_at 1446496_at | 2128 | 1.299 | 0.4132 | Yes | ||
| 36 | KIN | 1427317_at | 2140 | 1.293 | 0.4196 | Yes | ||
| 37 | SNRPD3 | 1422884_at 1422885_at 1449629_s_at | 2144 | 1.292 | 0.4264 | Yes | ||
| 38 | GEMIN6 | 1424300_at | 2236 | 1.252 | 0.4290 | Yes | ||
| 39 | SFRS11 | 1427269_at 1430077_at 1439095_at 1452371_at | 2384 | 1.197 | 0.4287 | Yes | ||
| 40 | SNRPB2 | 1426613_a_at 1452422_a_at | 2492 | 1.158 | 0.4300 | Yes | ||
| 41 | NOL3 | 1444786_at 1451503_at | 2555 | 1.141 | 0.4333 | Yes | ||
| 42 | AARS | 1423685_at 1451083_s_at | 2573 | 1.133 | 0.4386 | Yes | ||
| 43 | RBPMS | 1425652_s_at 1429359_s_at 1437161_x_at 1444765_at 1455936_a_at 1459078_at | 2741 | 1.086 | 0.4368 | Yes | ||
| 44 | DUSP11 | 1452594_at 1454785_at | 2770 | 1.081 | 0.4413 | Yes | ||
| 45 | NOLA2 | 1416605_at 1416606_s_at | 2933 | 1.039 | 0.4394 | Yes | ||
| 46 | SFRS2IP | 1434691_at 1443085_at 1452884_at 1452885_at 1460445_at | 2937 | 1.038 | 0.4449 | Yes | ||
| 47 | GEMIN5 | 1436310_at 1436311_at 1455453_at | 2992 | 1.025 | 0.4479 | Yes | ||
| 48 | U2AF1 | 1422509_at | 3069 | 1.010 | 0.4499 | Yes | ||
| 49 | SRRM1 | 1420934_a_at 1420935_a_at 1442330_at 1447447_s_at 1450045_at 1454689_at 1459828_at | 3295 | 0.966 | 0.4447 | Yes | ||
| 50 | HNRPF | 1450963_at | 3359 | 0.952 | 0.4469 | Yes | ||
| 51 | BCAS2 | 1418234_s_at 1437262_x_at 1458219_at | 3540 | 0.913 | 0.4436 | Yes | ||
| 52 | GRSF1 | 1433457_s_at 1433824_x_at 1437838_x_at 1439366_at | 3553 | 0.911 | 0.4479 | Yes | ||
| 53 | HNRPDL | 1420093_s_at 1420094_at 1424251_a_at 1424252_at 1428224_at 1428225_s_at 1449039_a_at 1456698_s_at | 3571 | 0.907 | 0.4520 | Yes | ||
| 54 | DHX38 | 1426786_s_at 1453751_at | 3667 | 0.884 | 0.4524 | Yes | ||
| 55 | CUGBP1 | 1423932_at 1425932_a_at 1426407_at 1426408_at 1427413_a_at | 3874 | 0.843 | 0.4475 | No | ||
| 56 | SFRS9 | 1417727_at | 3882 | 0.842 | 0.4517 | No | ||
| 57 | PTBP1 | 1424874_a_at 1450443_at 1458284_at | 4773 | 0.679 | 0.4145 | No | ||
| 58 | NCBP2 | 1423045_at 1423046_s_at 1450847_at | 4790 | 0.676 | 0.4174 | No | ||
| 59 | AGGF1 | 1425354_a_at | 5378 | 0.583 | 0.3936 | No | ||
| 60 | KHDRBS1 | 1418628_at 1418630_at | 5532 | 0.560 | 0.3896 | No | ||
| 61 | GEMIN7 | 1435764_a_at | 5952 | 0.507 | 0.3731 | No | ||
| 62 | RPS14 | 1436586_x_at | 5976 | 0.505 | 0.3748 | No | ||
| 63 | CDC2L1 | 1418841_s_at | 6131 | 0.484 | 0.3703 | No | ||
| 64 | PRPF18 | 1429614_at | 6166 | 0.481 | 0.3713 | No | ||
| 65 | PPAN | 1423703_at | 6791 | 0.409 | 0.3449 | No | ||
| 66 | HNRPD | 1425142_a_at 1446083_at | 7324 | 0.351 | 0.3224 | No | ||
| 67 | EXOSC2 | 1426630_at 1446994_at | 7606 | 0.321 | 0.3112 | No | ||
| 68 | METTL1 | 1419122_at 1439155_at 1447683_x_at | 7709 | 0.313 | 0.3082 | No | ||
| 69 | SF3A1 | 1419087_s_at 1445719_at 1446687_at 1449333_at | 7965 | 0.290 | 0.2980 | No | ||
| 70 | SF3B2 | 1429362_a_at 1433461_at 1438637_x_at 1453123_at 1456040_at 1456352_a_at 1459836_x_at | 8287 | 0.257 | 0.2847 | No | ||
| 71 | SFPQ | 1423795_at 1423796_at 1436898_at 1438458_a_at 1438459_x_at 1439058_at | 8629 | 0.223 | 0.2703 | No | ||
| 72 | EFTUD2 | 1416557_a_at 1438835_a_at 1438836_at 1456107_x_at | 8890 | 0.200 | 0.2594 | No | ||
| 73 | PPIG | 1434475_at 1435341_at 1436505_at | 8912 | 0.197 | 0.2595 | No | ||
| 74 | TXNL4A | 1419179_at | 8958 | 0.193 | 0.2585 | No | ||
| 75 | CPSF3 | 1422855_at 1437328_x_at 1437852_x_at | 9623 | 0.134 | 0.2287 | No | ||
| 76 | DDX39 | 1423643_at 1438168_x_at 1438287_x_at 1451065_a_at 1455814_x_at | 9785 | 0.119 | 0.2220 | No | ||
| 77 | LSM6 | 1431054_at 1437108_at 1437109_s_at 1455926_at | 9883 | 0.111 | 0.2181 | No | ||
| 78 | SF1 | 1422321_a_at 1423750_a_at 1423751_at 1459765_s_at 1459766_x_at | 9904 | 0.108 | 0.2178 | No | ||
| 79 | KHSRP | 1428174_x_at 1436813_x_at 1452690_at | 9965 | 0.104 | 0.2156 | No | ||
| 80 | SNRPD1 | 1416336_s_at | 10296 | 0.078 | 0.2009 | No | ||
| 81 | LSM5 | 1418656_at | 10591 | 0.053 | 0.1877 | No | ||
| 82 | RBMY1A1 | 1420786_a_at | 10808 | 0.035 | 0.1779 | No | ||
| 83 | INTS6 | 1423274_at 1423275_at 1436089_at 1443128_at | 10862 | 0.031 | 0.1757 | No | ||
| 84 | CSTF3 | 1424723_s_at 1443909_at | 11465 | -0.025 | 0.1482 | No | ||
| 85 | RNMT | 1436328_at 1440002_at 1446442_at 1453106_a_at | 11665 | -0.044 | 0.1393 | No | ||
| 86 | USP39 | 1437007_x_at 1460209_at | 11889 | -0.062 | 0.1294 | No | ||
| 87 | UPF1 | 1419685_at | 12500 | -0.118 | 0.1020 | No | ||
| 88 | RBM5 | 1438069_a_at 1441319_at 1444618_at 1444971_at 1452186_at 1452187_at 1456262_at | 12628 | -0.128 | 0.0969 | No | ||
| 89 | WDR57 | 1428264_at 1452713_a_at | 12904 | -0.155 | 0.0851 | No | ||
| 90 | SRPK2 | 1417134_at 1417135_at 1417136_s_at 1431372_at 1435746_at 1440080_at 1442241_at 1442980_at 1447755_at 1448603_at | 13008 | -0.165 | 0.0813 | No | ||
| 91 | HNRPUL1 | 1451984_at | 13037 | -0.168 | 0.0809 | No | ||
| 92 | PRPF31 | 1448633_at 1453005_at 1457083_at | 13078 | -0.171 | 0.0800 | No | ||
| 93 | LSM4 | 1448622_at | 13372 | -0.197 | 0.0676 | No | ||
| 94 | CPSF1 | 1417665_a_at | 13443 | -0.203 | 0.0654 | No | ||
| 95 | BICD1 | 1427530_at 1437548_at 1438701_at 1451684_a_at | 13886 | -0.244 | 0.0465 | No | ||
| 96 | ZNF638 | 1417791_a_at 1417792_at 1447983_at 1458523_at | 14045 | -0.261 | 0.0406 | No | ||
| 97 | RBM6 | 1417213_a_at 1440223_at 1452466_a_at 1458912_at | 14430 | -0.299 | 0.0246 | No | ||
| 98 | SNRPF | 1428672_at | 14478 | -0.305 | 0.0241 | No | ||
| 99 | SRPK1 | 1454042_a_at 1455107_at | 14582 | -0.315 | 0.0211 | No | ||
| 100 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 14703 | -0.329 | 0.0173 | No | ||
| 101 | LSM3 | 1448536_at | 14770 | -0.337 | 0.0161 | No | ||
| 102 | PPARGC1A | 1434099_at 1434100_x_at 1437751_at 1456395_at 1460336_at | 14869 | -0.348 | 0.0135 | No | ||
| 103 | ADAT1 | 1420748_a_at 1425390_at 1444049_at | 14938 | -0.355 | 0.0123 | No | ||
| 104 | SNW1 | 1429002_at 1429003_at 1459026_at | 15023 | -0.365 | 0.0104 | No | ||
| 105 | PRPF8 | 1422453_at | 15399 | -0.406 | -0.0046 | No | ||
| 106 | CDC40 | 1442667_at 1445348_at 1453342_at 1457366_at | 15645 | -0.436 | -0.0135 | No | ||
| 107 | SARS | 1426257_a_at 1452000_s_at | 16324 | -0.522 | -0.0419 | No | ||
| 108 | ASCC3L1 | 1454772_at 1460552_at | 17034 | -0.626 | -0.0710 | No | ||
| 109 | SPOP | 1416524_at 1416525_at 1444578_at 1447827_x_at 1455510_at 1458886_at | 17100 | -0.636 | -0.0706 | No | ||
| 110 | INTS10 | 1439712_at | 17159 | -0.647 | -0.0698 | No | ||
| 111 | DBR1 | 1451641_at | 17344 | -0.677 | -0.0746 | No | ||
| 112 | TRNT1 | 1425561_at 1425562_s_at | 17452 | -0.695 | -0.0758 | No | ||
| 113 | INTS5 | 1417887_at 1434011_a_at 1434012_at | 17584 | -0.718 | -0.0779 | No | ||
| 114 | LSM1 | 1423873_at 1441452_at | 17634 | -0.728 | -0.0762 | No | ||
| 115 | DHX16 | 1423925_at | 17874 | -0.776 | -0.0830 | No | ||
| 116 | SLBP | 1430058_at 1460168_at | 18125 | -0.829 | -0.0901 | No | ||
| 117 | FBL | 1416684_at 1416685_s_at | 18145 | -0.833 | -0.0864 | No | ||
| 118 | ZNF346 | 1417088_at 1448581_at | 18212 | -0.853 | -0.0849 | No | ||
| 119 | SIP1 | 1451044_at | 18310 | -0.876 | -0.0846 | No | ||
| 120 | SNRP70 | 1429009_at 1429010_at 1429011_x_at 1451104_a_at 1457047_at | 18499 | -0.920 | -0.0883 | No | ||
| 121 | DDX54 | 1438853_x_at 1447882_x_at 1460396_at | 18527 | -0.926 | -0.0846 | No | ||
| 122 | FARS2 | 1431354_a_at 1439406_x_at | 18546 | -0.933 | -0.0804 | No | ||
| 123 | SFRS7 | 1424033_at 1424883_s_at 1436871_at | 18695 | -0.964 | -0.0820 | No | ||
| 124 | DHX8 | 1426629_at | 18696 | -0.965 | -0.0768 | No | ||
| 125 | CSTF1 | 1417117_at 1448597_at | 18947 | -1.008 | -0.0829 | No | ||
| 126 | SFRS4 | 1448778_at | 18986 | -1.014 | -0.0792 | No | ||
| 127 | RNPS1 | 1437027_x_at 1437359_at 1437851_x_at 1438267_x_at 1448324_at | 19075 | -1.028 | -0.0777 | No | ||
| 128 | RBM3 | 1429169_at | 19113 | -1.034 | -0.0738 | No | ||
| 129 | GEMIN4 | 1433622_at | 19225 | -1.058 | -0.0732 | No | ||
| 130 | SNRPN | 1415895_at 1415896_x_at 1432096_at 1435716_x_at | 19313 | -1.082 | -0.0714 | No | ||
| 131 | SNRPB | 1419260_a_at 1437193_s_at | 19808 | -1.239 | -0.0874 | No | ||
| 132 | RBMS2 | 1423540_at 1433979_at | 19873 | -1.261 | -0.0836 | No | ||
| 133 | SFRS8 | 1424997_at 1433677_at 1434966_at 1438674_a_at 1438675_at 1443905_at 1454683_at 1456880_at | 19889 | -1.268 | -0.0775 | No | ||
| 134 | DDX23 | 1430050_at | 20026 | -1.330 | -0.0765 | No | ||
| 135 | RBMS1 | 1418703_at 1434005_at 1441437_at 1458578_at 1459968_at | 20132 | -1.382 | -0.0739 | No | ||
| 136 | CPSF3L | 1423964_at | 20406 | -1.515 | -0.0783 | No | ||
| 137 | SNRPD2 | 1452680_at | 20486 | -1.550 | -0.0736 | No | ||
| 138 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 20908 | -1.849 | -0.0830 | No | ||
| 139 | EXOSC7 | 1448365_at | 20935 | -1.866 | -0.0742 | No | ||
| 140 | SF3B3 | 1415985_at 1430075_at | 21067 | -1.964 | -0.0696 | No | ||
| 141 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 21222 | -2.084 | -0.0655 | No | ||
| 142 | PRPF3 | 1424314_at 1457390_at | 21533 | -2.495 | -0.0663 | No | ||
| 143 | INTS3 | 1423921_at 1423922_s_at | 21908 | -7.184 | -0.0449 | No | ||
| 144 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 21936 | -8.578 | -0.0000 | No |