Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
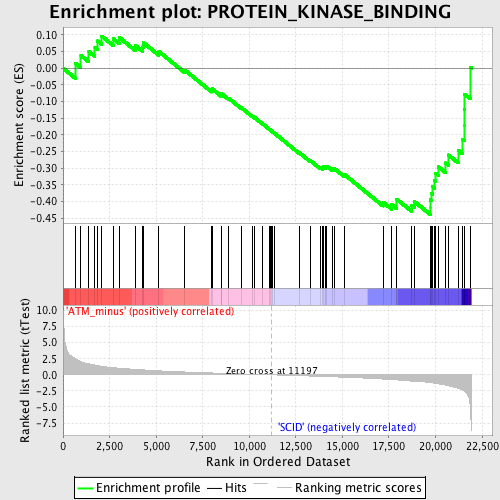
| GeneSet | PROTEIN_KINASE_BINDING |
| Enrichment Score (ES) | -0.43890575 |
| Normalized Enrichment Score (NES) | -1.5316488 |
| Nominal p-value | 0.025462963 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
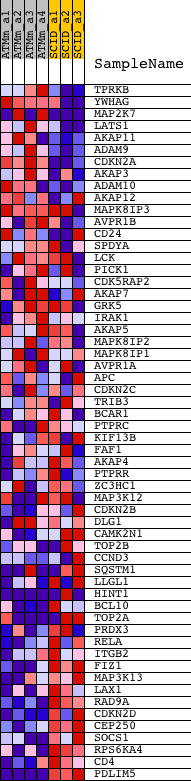
| 1 | TPRKB | 1424992_at 1425410_at 1425682_a_at | 663 | 2.453 | 0.0147 | No | ||
| 2 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 958 | 2.020 | 0.0383 | No | ||
| 3 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 1386 | 1.693 | 0.0499 | No | ||
| 4 | LATS1 | 1427679_at 1460264_at | 1710 | 1.501 | 0.0626 | No | ||
| 5 | AKAP11 | 1434764_at | 1862 | 1.427 | 0.0819 | No | ||
| 6 | ADAM9 | 1416094_at | 2075 | 1.321 | 0.0965 | No | ||
| 7 | CDKN2A | 1450140_a_at | 2688 | 1.102 | 0.0887 | No | ||
| 8 | AKAP3 | 1421531_at 1456591_x_at | 3027 | 1.018 | 0.0920 | No | ||
| 9 | ADAM10 | 1450104_at 1460083_at | 3870 | 0.844 | 0.0690 | No | ||
| 10 | AKAP12 | 1419706_a_at 1458235_at | 4288 | 0.766 | 0.0640 | No | ||
| 11 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 4327 | 0.761 | 0.0762 | No | ||
| 12 | AVPR1B | 1422204_at | 5145 | 0.616 | 0.0501 | No | ||
| 13 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 6541 | 0.437 | -0.0056 | No | ||
| 14 | SPDYA | 1429470_at 1432134_at 1441390_at | 7979 | 0.289 | -0.0660 | No | ||
| 15 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 8004 | 0.287 | -0.0618 | No | ||
| 16 | PICK1 | 1419384_at | 8497 | 0.235 | -0.0800 | No | ||
| 17 | CDK5RAP2 | 1443976_at 1447103_at 1452618_at | 8514 | 0.233 | -0.0764 | No | ||
| 18 | AKAP7 | 1422143_at 1433905_at | 8897 | 0.200 | -0.0902 | No | ||
| 19 | GRK5 | 1446361_at 1446998_at 1449514_at | 9571 | 0.139 | -0.1185 | No | ||
| 20 | IRAK1 | 1448668_a_at 1460649_at | 10167 | 0.089 | -0.1440 | No | ||
| 21 | AKAP5 | 1447301_at | 10302 | 0.077 | -0.1487 | No | ||
| 22 | MAPK8IP2 | 1418785_at 1418786_at 1435045_s_at 1449225_a_at 1455194_at | 10714 | 0.043 | -0.1667 | No | ||
| 23 | MAPK8IP1 | 1425679_a_at 1440619_at | 11065 | 0.011 | -0.1825 | No | ||
| 24 | AVPR1A | 1418603_at 1418604_at | 11156 | 0.003 | -0.1866 | No | ||
| 25 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 11193 | 0.000 | -0.1882 | No | ||
| 26 | CDKN2C | 1416868_at 1439164_at | 11228 | -0.002 | -0.1897 | No | ||
| 27 | TRIB3 | 1426065_a_at 1456225_x_at | 11356 | -0.015 | -0.1952 | No | ||
| 28 | BCAR1 | 1439388_s_at 1450622_at | 12678 | -0.132 | -0.2532 | No | ||
| 29 | PTPRC | 1422124_a_at 1440165_at | 13287 | -0.189 | -0.2775 | No | ||
| 30 | KIF13B | 1433158_at 1433159_at 1436056_at 1453276_at | 13808 | -0.236 | -0.2970 | No | ||
| 31 | FAF1 | 1418242_at 1434370_s_at | 13938 | -0.249 | -0.2983 | No | ||
| 32 | AKAP4 | 1419019_a_at 1431868_at 1451661_at 1451662_x_at | 13965 | -0.252 | -0.2948 | No | ||
| 33 | PTPRR | 1426047_a_at | 14115 | -0.268 | -0.2967 | No | ||
| 34 | ZC3HC1 | 1426654_at | 14152 | -0.272 | -0.2934 | No | ||
| 35 | MAP3K12 | 1417115_at 1438908_at 1456565_s_at | 14452 | -0.301 | -0.3015 | No | ||
| 36 | CDKN2B | 1449152_at | 14572 | -0.314 | -0.3012 | No | ||
| 37 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 15106 | -0.374 | -0.3187 | No | ||
| 38 | CAMK2N1 | 1456609_at | 17180 | -0.651 | -0.4015 | No | ||
| 39 | TOP2B | 1416731_at 1416732_at 1448458_at 1456688_at | 17652 | -0.731 | -0.4096 | No | ||
| 40 | CCND3 | 1415907_at 1437584_at 1444323_at 1446567_at 1447434_at 1457549_at | 17921 | -0.785 | -0.4075 | No | ||
| 41 | SQSTM1 | 1440076_at 1442237_at 1444021_at 1450957_a_at | 17923 | -0.785 | -0.3931 | No | ||
| 42 | LLGL1 | 1416621_at | 18709 | -0.967 | -0.4113 | No | ||
| 43 | HINT1 | 1424017_a_at 1424018_at 1457286_at | 18873 | -0.997 | -0.4004 | No | ||
| 44 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 19716 | -1.203 | -0.4168 | Yes | ||
| 45 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 19718 | -1.203 | -0.3948 | Yes | ||
| 46 | PRDX3 | 1416292_at | 19765 | -1.219 | -0.3745 | Yes | ||
| 47 | RELA | 1419536_a_at | 19848 | -1.253 | -0.3553 | Yes | ||
| 48 | ITGB2 | 1450678_at | 19948 | -1.288 | -0.3362 | Yes | ||
| 49 | FIZ1 | 1423733_a_at | 20003 | -1.316 | -0.3145 | Yes | ||
| 50 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 20161 | -1.392 | -0.2962 | Yes | ||
| 51 | LAX1 | 1438687_at | 20539 | -1.588 | -0.2842 | Yes | ||
| 52 | RAD9A | 1418404_at | 20695 | -1.692 | -0.2603 | Yes | ||
| 53 | CDKN2D | 1416253_at | 21221 | -2.083 | -0.2461 | Yes | ||
| 54 | CEP250 | 1421512_at | 21460 | -2.403 | -0.2129 | Yes | ||
| 55 | SOCS1 | 1440047_at 1450446_a_at | 21547 | -2.517 | -0.1706 | Yes | ||
| 56 | RPS6KA4 | 1438243_at 1448498_at | 21550 | -2.522 | -0.1244 | Yes | ||
| 57 | CD4 | 1419696_at 1427779_a_at | 21569 | -2.571 | -0.0781 | Yes | ||
| 58 | PDLIM5 | 1429783_at 1438186_at 1442710_at 1446339_at | 21872 | -5.168 | 0.0029 | Yes |