Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
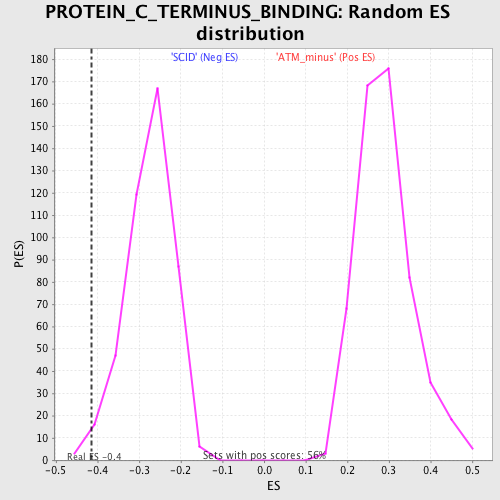
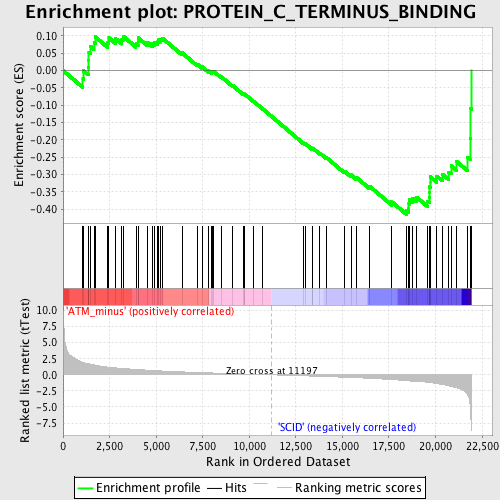
| GeneSet | PROTEIN_C_TERMINUS_BINDING |
| Enrichment Score (ES) | -0.41490433 |
| Normalized Enrichment Score (NES) | -1.5071193 |
| Nominal p-value | 0.020224718 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

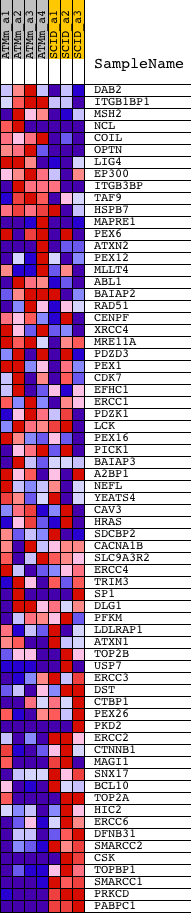
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DAB2 | 1420498_a_at 1423805_at 1429693_at 1430604_a_at | 1066 | 1.907 | -0.0240 | No | ||
| 2 | ITGB1BP1 | 1422157_a_at 1445697_at | 1088 | 1.884 | -0.0005 | No | ||
| 3 | MSH2 | 1416988_at 1447888_x_at | 1351 | 1.709 | 0.0097 | No | ||
| 4 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 1383 | 1.694 | 0.0302 | No | ||
| 5 | COIL | 1416692_at | 1387 | 1.692 | 0.0520 | No | ||
| 6 | OPTN | 1429778_at 1435679_at | 1463 | 1.653 | 0.0700 | No | ||
| 7 | LIG4 | 1439487_at | 1659 | 1.532 | 0.0810 | No | ||
| 8 | EP300 | 1434765_at | 1726 | 1.492 | 0.0973 | No | ||
| 9 | ITGB3BP | 1420665_at 1442993_at 1449953_at 1456097_a_at | 2399 | 1.191 | 0.0820 | No | ||
| 10 | TAF9 | 1422778_at 1451509_at | 2462 | 1.167 | 0.0943 | No | ||
| 11 | HSPB7 | 1421289_at 1421290_at 1434927_at | 2802 | 1.072 | 0.0927 | No | ||
| 12 | MAPRE1 | 1422764_at 1422765_at 1428819_at 1428820_at 1450740_a_at | 3155 | 0.995 | 0.0895 | No | ||
| 13 | PEX6 | 1424078_s_at 1438138_a_at 1451226_at 1454738_x_at 1456302_at | 3219 | 0.983 | 0.0994 | No | ||
| 14 | ATXN2 | 1419866_s_at 1438143_s_at 1438144_x_at 1443516_at 1459363_at 1460653_at | 3914 | 0.836 | 0.0785 | No | ||
| 15 | PEX12 | 1416259_at | 4029 | 0.814 | 0.0839 | No | ||
| 16 | MLLT4 | 1431799_at 1436303_at | 4034 | 0.813 | 0.0942 | No | ||
| 17 | ABL1 | 1423999_at 1441291_at 1444134_at 1445153_at | 4519 | 0.722 | 0.0814 | No | ||
| 18 | BAIAP2 | 1425656_a_at 1435128_at 1451027_at 1451028_at | 4795 | 0.675 | 0.0776 | No | ||
| 19 | RAD51 | 1418281_at | 4919 | 0.654 | 0.0805 | No | ||
| 20 | CENPF | 1427161_at 1452334_at 1458447_at | 5054 | 0.631 | 0.0825 | No | ||
| 21 | XRCC4 | 1424601_at 1424602_s_at 1445922_at | 5101 | 0.623 | 0.0885 | No | ||
| 22 | MRE11A | 1416748_a_at 1441990_at 1458509_at | 5237 | 0.602 | 0.0901 | No | ||
| 23 | PDZD3 | 1449330_at | 5331 | 0.589 | 0.0935 | No | ||
| 24 | PEX1 | 1428716_at 1440267_at | 6388 | 0.453 | 0.0511 | No | ||
| 25 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 7205 | 0.362 | 0.0185 | No | ||
| 26 | EFHC1 | 1453159_at | 7460 | 0.337 | 0.0113 | No | ||
| 27 | ERCC1 | 1417328_at 1437447_s_at 1441838_at 1445266_at | 7810 | 0.303 | -0.0008 | No | ||
| 28 | PDZK1 | 1431701_a_at | 7970 | 0.289 | -0.0043 | No | ||
| 29 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 8004 | 0.287 | -0.0021 | No | ||
| 30 | PEX16 | 1425021_a_at 1436253_at 1447801_x_at | 8080 | 0.278 | -0.0019 | No | ||
| 31 | PICK1 | 1419384_at | 8497 | 0.235 | -0.0179 | No | ||
| 32 | BAIAP3 | 1427509_at | 9115 | 0.180 | -0.0437 | No | ||
| 33 | A2BP1 | 1418314_a_at 1438217_at 1447735_x_at 1455358_at 1456921_at | 9663 | 0.130 | -0.0671 | No | ||
| 34 | NEFL | 1426255_at | 9738 | 0.124 | -0.0689 | No | ||
| 35 | YEATS4 | 1423105_a_at | 9741 | 0.124 | -0.0673 | No | ||
| 36 | CAV3 | 1418413_at | 10228 | 0.084 | -0.0885 | No | ||
| 37 | HRAS | 1422407_s_at 1424132_at | 10720 | 0.043 | -0.1104 | No | ||
| 38 | SDCBP2 | 1424090_at | 12915 | -0.156 | -0.2087 | No | ||
| 39 | CACNA1B | 1425812_a_at 1436602_x_at 1439612_at 1460608_at | 12991 | -0.163 | -0.2100 | No | ||
| 40 | SLC9A3R2 | 1428954_at 1428955_x_at 1431208_a_at 1439368_a_at 1439369_x_at 1452976_a_at | 13388 | -0.199 | -0.2255 | No | ||
| 41 | ERCC4 | 1432272_a_at 1440237_at 1441003_at 1448984_at | 13394 | -0.199 | -0.2232 | No | ||
| 42 | TRIM3 | 1451663_a_at | 13795 | -0.234 | -0.2384 | No | ||
| 43 | SP1 | 1418180_at 1448994_at 1454852_at | 14131 | -0.269 | -0.2502 | No | ||
| 44 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 15106 | -0.374 | -0.2899 | No | ||
| 45 | PFKM | 1416780_at | 15461 | -0.413 | -0.3008 | No | ||
| 46 | LDLRAP1 | 1424378_at 1457162_at | 15742 | -0.447 | -0.3078 | No | ||
| 47 | ATXN1 | 1438294_at 1445695_at 1450499_at 1457605_at | 16478 | -0.542 | -0.3343 | No | ||
| 48 | TOP2B | 1416731_at 1416732_at 1448458_at 1456688_at | 17652 | -0.731 | -0.3785 | No | ||
| 49 | USP7 | 1419920_s_at 1419921_s_at 1434371_x_at 1437118_at 1438249_at 1454948_at 1454949_at | 18449 | -0.908 | -0.4031 | Yes | ||
| 50 | ERCC3 | 1448497_at | 18568 | -0.937 | -0.3964 | Yes | ||
| 51 | DST | 1421117_at 1421276_a_at 1423626_at 1442395_at 1446943_at 1450119_at 1458075_at 1459098_at 1459356_at | 18573 | -0.938 | -0.3844 | Yes | ||
| 52 | CTBP1 | 1415702_a_at | 18588 | -0.942 | -0.3728 | Yes | ||
| 53 | PEX26 | 1451393_at | 18749 | -0.974 | -0.3675 | Yes | ||
| 54 | PKD2 | 1417753_at 1441870_s_at | 18989 | -1.014 | -0.3653 | Yes | ||
| 55 | ERCC2 | 1460727_at | 19582 | -1.158 | -0.3773 | Yes | ||
| 56 | CTNNB1 | 1420811_a_at 1430533_a_at 1450008_a_at | 19662 | -1.184 | -0.3656 | Yes | ||
| 57 | MAGI1 | 1427246_at 1440071_at 1440871_at 1443231_at 1445088_at 1445952_at 1446050_at 1451893_s_at 1452369_at 1457572_at 1458240_at | 19672 | -1.188 | -0.3506 | Yes | ||
| 58 | SNX17 | 1455955_s_at | 19679 | -1.190 | -0.3354 | Yes | ||
| 59 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 19716 | -1.203 | -0.3215 | Yes | ||
| 60 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 19718 | -1.203 | -0.3059 | Yes | ||
| 61 | HIC2 | 1427581_at 1437571_at 1454944_at | 20070 | -1.354 | -0.3044 | Yes | ||
| 62 | ERCC6 | 1441595_at 1442604_at | 20368 | -1.495 | -0.2986 | Yes | ||
| 63 | DFNB31 | 1432555_at 1436485_s_at 1436486_x_at 1442507_at 1447438_at | 20718 | -1.706 | -0.2924 | Yes | ||
| 64 | SMARCC2 | 1428382_at | 20836 | -1.782 | -0.2747 | Yes | ||
| 65 | CSK | 1423518_at 1439744_at | 21126 | -2.013 | -0.2618 | Yes | ||
| 66 | TOPBP1 | 1452241_at 1458480_at | 21726 | -3.079 | -0.2492 | Yes | ||
| 67 | SMARCC1 | 1423416_at 1423417_at 1455246_at 1459824_at | 21859 | -4.672 | -0.1947 | Yes | ||
| 68 | PRKCD | 1422847_a_at 1442256_at | 21900 | -6.694 | -0.1097 | Yes | ||
| 69 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 21936 | -8.578 | -0.0000 | Yes |