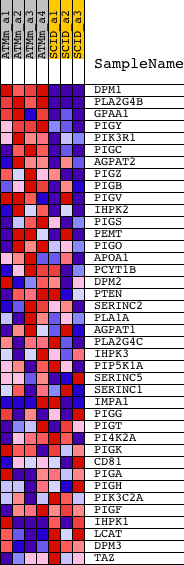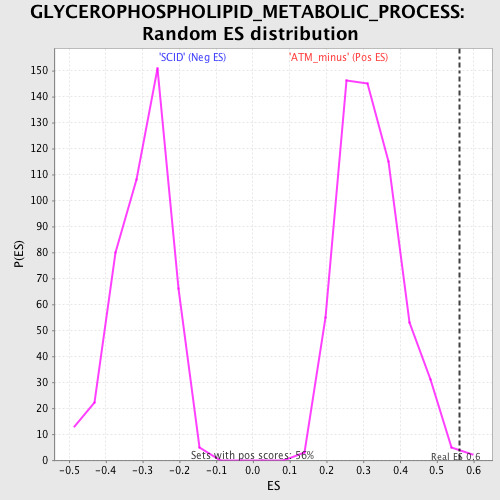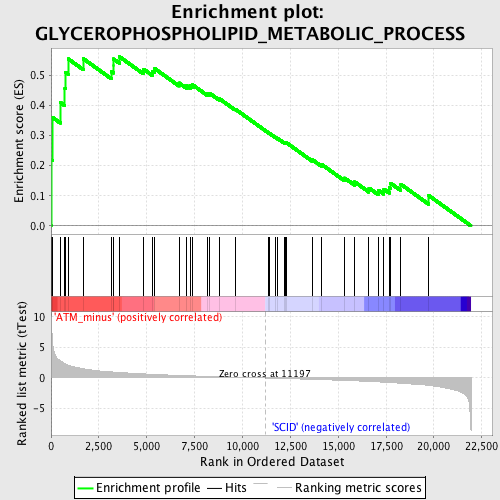
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | GLYCEROPHOSPHOLIPID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.56155604 |
| Normalized Enrichment Score (NES) | 1.7642251 |
| Nominal p-value | 0.0036036037 |
| FDR q-value | 0.41694897 |
| FWER p-Value | 0.894 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DPM1 | 1419353_at 1449438_at | 8 | 9.129 | 0.2199 | Yes | ||
| 2 | PLA2G4B | 1425045_at 1456047_at | 69 | 5.936 | 0.3604 | Yes | ||
| 3 | GPAA1 | 1438152_at 1448225_at 1455954_x_at | 482 | 2.837 | 0.4100 | Yes | ||
| 4 | PIGY | 1428556_at | 720 | 2.358 | 0.4561 | Yes | ||
| 5 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 749 | 2.311 | 0.5105 | Yes | ||
| 6 | PIGC | 1424189_at 1424190_at | 893 | 2.104 | 0.5548 | Yes | ||
| 7 | AGPAT2 | 1428821_at | 1686 | 1.518 | 0.5552 | Yes | ||
| 8 | PIGZ | 1440143_at 1441933_x_at 1458306_at | 3157 | 0.995 | 0.5121 | Yes | ||
| 9 | PIGB | 1420128_s_at 1421257_at 1435958_at | 3253 | 0.973 | 0.5312 | Yes | ||
| 10 | PIGV | 1437663_at 1455075_at | 3255 | 0.973 | 0.5546 | Yes | ||
| 11 | IHPK2 | 1428373_at 1435319_at | 3582 | 0.903 | 0.5616 | Yes | ||
| 12 | PIGS | 1433429_at | 4826 | 0.670 | 0.5210 | No | ||
| 13 | PEMT | 1450612_a_at | 5283 | 0.596 | 0.5145 | No | ||
| 14 | PIGO | 1415992_at 1437142_a_at 1438155_x_at | 5417 | 0.577 | 0.5224 | No | ||
| 15 | APOA1 | 1419232_a_at 1419233_x_at 1438840_x_at 1455201_x_at | 6682 | 0.420 | 0.4748 | No | ||
| 16 | PCYT1B | 1436124_at 1437648_at | 7088 | 0.376 | 0.4654 | No | ||
| 17 | DPM2 | 1415675_at | 7275 | 0.356 | 0.4655 | No | ||
| 18 | PTEN | 1422553_at 1441593_at 1444325_at 1450655_at 1454722_at 1455728_at 1457493_at | 7387 | 0.345 | 0.4687 | No | ||
| 19 | SERINC2 | 1434382_at | 8161 | 0.270 | 0.4399 | No | ||
| 20 | PLA1A | 1417785_at | 8285 | 0.258 | 0.4405 | No | ||
| 21 | AGPAT1 | 1421024_at 1421025_at | 8784 | 0.210 | 0.4228 | No | ||
| 22 | PLA2G4C | 1436712_at 1444662_at | 9615 | 0.135 | 0.3882 | No | ||
| 23 | IHPK3 | 1443503_at | 11349 | -0.014 | 0.3094 | No | ||
| 24 | PIP5K1A | 1421833_at 1421834_at 1450389_s_at | 11427 | -0.022 | 0.3064 | No | ||
| 25 | SERINC5 | 1433571_at | 11714 | -0.047 | 0.2945 | No | ||
| 26 | SERINC1 | 1415838_at 1437513_a_at 1448108_at 1454811_a_at | 11849 | -0.059 | 0.2898 | No | ||
| 27 | IMPA1 | 1423127_at 1430495_at 1436848_x_at | 12207 | -0.090 | 0.2756 | No | ||
| 28 | PIGG | 1444640_at | 12228 | -0.092 | 0.2769 | No | ||
| 29 | PIGT | 1428740_a_at 1437594_x_at 1447382_at | 12273 | -0.096 | 0.2772 | No | ||
| 30 | PI4K2A | 1433462_a_at 1454605_a_at | 13643 | -0.220 | 0.2200 | No | ||
| 31 | PIGK | 1428488_at 1435352_at 1452801_at | 14119 | -0.269 | 0.2048 | No | ||
| 32 | CD81 | 1416330_at | 15333 | -0.398 | 0.1590 | No | ||
| 33 | PIGA | 1427305_at 1453497_a_at | 15863 | -0.461 | 0.1460 | No | ||
| 34 | PIGH | 1430589_at 1455860_at | 16604 | -0.562 | 0.1257 | No | ||
| 35 | PIK3C2A | 1421023_at 1425862_a_at 1443655_s_at | 17090 | -0.634 | 0.1189 | No | ||
| 36 | PIGF | 1448896_at | 17384 | -0.683 | 0.1220 | No | ||
| 37 | IHPK1 | 1422968_at 1422969_s_at 1438845_at 1438846_x_at 1452694_at | 17671 | -0.734 | 0.1266 | No | ||
| 38 | LCAT | 1417043_at | 17728 | -0.744 | 0.1420 | No | ||
| 39 | DPM3 | 1452729_at | 18274 | -0.866 | 0.1380 | No | ||
| 40 | TAZ | 1416207_at 1453785_at | 19731 | -1.209 | 0.1007 | No |