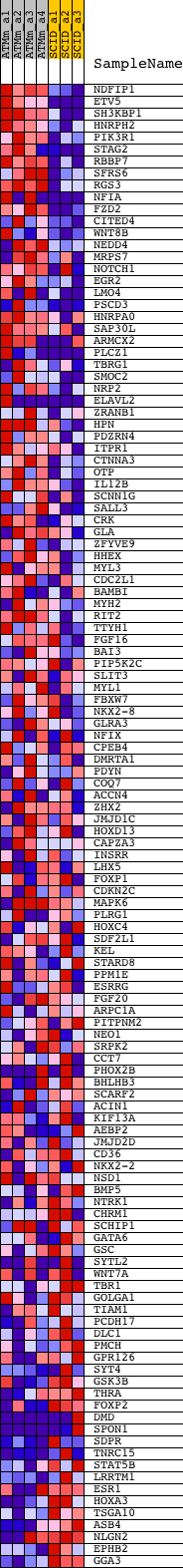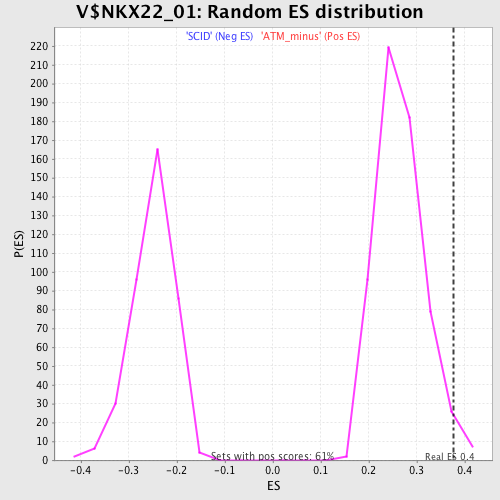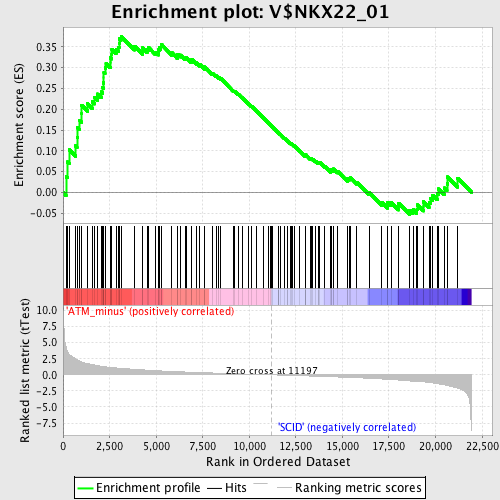
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | V$NKX22_01 |
| Enrichment Score (ES) | 0.3759129 |
| Normalized Enrichment Score (NES) | 1.4172016 |
| Nominal p-value | 0.018003274 |
| FDR q-value | 0.957442 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NDFIP1 | 1424820_a_at 1424821_at 1451493_at | 160 | 4.284 | 0.0378 | Yes | ||
| 2 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 226 | 3.659 | 0.0734 | Yes | ||
| 3 | SH3KBP1 | 1431592_a_at 1432269_a_at 1446562_at 1456261_at 1460337_at | 326 | 3.189 | 0.1025 | Yes | ||
| 4 | HNRPH2 | 1415963_at | 676 | 2.426 | 0.1121 | Yes | ||
| 5 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 749 | 2.311 | 0.1331 | Yes | ||
| 6 | STAG2 | 1421849_at 1450396_at | 783 | 2.268 | 0.1555 | Yes | ||
| 7 | RBBP7 | 1415775_at 1456227_x_at | 895 | 2.102 | 0.1726 | Yes | ||
| 8 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 990 | 1.984 | 0.1892 | Yes | ||
| 9 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 998 | 1.979 | 0.2098 | Yes | ||
| 10 | NFIA | 1421162_a_at 1421163_a_at 1427733_a_at 1438236_at 1440660_at 1441547_at 1446742_at 1446990_at 1456087_at 1459431_at | 1316 | 1.727 | 0.2134 | Yes | ||
| 11 | FZD2 | 1418532_at 1418533_s_at | 1567 | 1.586 | 0.2187 | Yes | ||
| 12 | CITED4 | 1425400_a_at 1438261_at | 1700 | 1.508 | 0.2285 | Yes | ||
| 13 | WNT8B | 1421439_at | 1851 | 1.434 | 0.2368 | Yes | ||
| 14 | NEDD4 | 1421955_a_at 1444044_at 1450431_a_at 1451109_a_at | 2046 | 1.334 | 0.2419 | Yes | ||
| 15 | MRPS7 | 1453725_a_at | 2125 | 1.300 | 0.2521 | Yes | ||
| 16 | NOTCH1 | 1418633_at 1418634_at | 2169 | 1.283 | 0.2636 | Yes | ||
| 17 | EGR2 | 1427682_a_at 1427683_at | 2191 | 1.273 | 0.2761 | Yes | ||
| 18 | LMO4 | 1420981_a_at 1439651_at | 2192 | 1.273 | 0.2895 | Yes | ||
| 19 | PSCD3 | 1418758_a_at 1431707_a_at 1446268_at | 2252 | 1.245 | 0.2999 | Yes | ||
| 20 | HNRPA0 | 1428407_at | 2302 | 1.221 | 0.3105 | Yes | ||
| 21 | SAP30L | 1415718_at 1447379_at 1460071_at | 2537 | 1.146 | 0.3119 | Yes | ||
| 22 | ARMCX2 | 1415780_a_at 1437849_x_at 1456739_x_at | 2539 | 1.146 | 0.3239 | Yes | ||
| 23 | PLCZ1 | 1422658_at 1432405_a_at | 2609 | 1.124 | 0.3326 | Yes | ||
| 24 | TBRG1 | 1452648_at | 2618 | 1.122 | 0.3441 | Yes | ||
| 25 | SMOC2 | 1415935_at 1431362_a_at | 2846 | 1.061 | 0.3449 | Yes | ||
| 26 | NRP2 | 1425433_a_at 1426528_at 1435349_at 1452552_at 1456778_at 1459256_at | 2978 | 1.029 | 0.3497 | Yes | ||
| 27 | ELAVL2 | 1421881_a_at 1421882_a_at 1421883_at 1433052_at | 3008 | 1.022 | 0.3591 | Yes | ||
| 28 | ZRANB1 | 1415712_at 1427200_at 1436228_at | 3020 | 1.019 | 0.3694 | Yes | ||
| 29 | HPN | 1420712_a_at | 3109 | 1.002 | 0.3759 | Yes | ||
| 30 | PDZRN4 | 1456512_at | 3840 | 0.850 | 0.3514 | No | ||
| 31 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 4275 | 0.769 | 0.3396 | No | ||
| 32 | CTNNA3 | 1441560_at | 4277 | 0.768 | 0.3477 | No | ||
| 33 | OTP | 1422285_at | 4511 | 0.723 | 0.3446 | No | ||
| 34 | IL12B | 1419530_at 1449497_at | 4597 | 0.707 | 0.3482 | No | ||
| 35 | SCNN1G | 1419079_at | 4982 | 0.643 | 0.3373 | No | ||
| 36 | SALL3 | 1441186_at 1452386_at | 5133 | 0.618 | 0.3370 | No | ||
| 37 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 5140 | 0.617 | 0.3432 | No | ||
| 38 | GLA | 1418248_at 1449006_at | 5166 | 0.612 | 0.3485 | No | ||
| 39 | ZFYVE9 | 1440348_at | 5257 | 0.599 | 0.3507 | No | ||
| 40 | HHEX | 1423319_at | 5271 | 0.597 | 0.3564 | No | ||
| 41 | MYL3 | 1427768_s_at 1427769_x_at 1428266_at | 5846 | 0.519 | 0.3356 | No | ||
| 42 | CDC2L1 | 1418841_s_at | 6131 | 0.484 | 0.3276 | No | ||
| 43 | BAMBI | 1423753_at | 6155 | 0.482 | 0.3317 | No | ||
| 44 | MYH2 | 1425153_at | 6288 | 0.465 | 0.3305 | No | ||
| 45 | RIT2 | 1421779_at 1448125_at 1460166_at | 6549 | 0.436 | 0.3232 | No | ||
| 46 | TTYH1 | 1422694_at 1422696_at 1426617_a_at | 6622 | 0.427 | 0.3244 | No | ||
| 47 | FGF16 | 1420806_at | 6902 | 0.397 | 0.3158 | No | ||
| 48 | BAI3 | 1446358_at 1454782_at | 6913 | 0.395 | 0.3195 | No | ||
| 49 | PIP5K2C | 1416387_at 1416388_at | 7166 | 0.368 | 0.3118 | No | ||
| 50 | SLIT3 | 1427086_at 1452296_at | 7345 | 0.349 | 0.3073 | No | ||
| 51 | MYL1 | 1452651_a_at | 7591 | 0.323 | 0.2995 | No | ||
| 52 | FBXW7 | 1424986_s_at 1451558_at | 7612 | 0.321 | 0.3020 | No | ||
| 53 | NKX2-8 | 1422284_at | 8019 | 0.285 | 0.2864 | No | ||
| 54 | GLRA3 | 1450239_at | 8210 | 0.265 | 0.2805 | No | ||
| 55 | NFIX | 1423493_a_at 1436363_a_at 1436364_x_at 1451443_at 1459909_at | 8342 | 0.251 | 0.2771 | No | ||
| 56 | CPEB4 | 1420617_at 1420618_at 1449931_at | 8465 | 0.238 | 0.2740 | No | ||
| 57 | DMRTA1 | 1441579_at | 9162 | 0.176 | 0.2440 | No | ||
| 58 | PDYN | 1416266_at | 9210 | 0.172 | 0.2436 | No | ||
| 59 | COQ7 | 1416665_at | 9400 | 0.154 | 0.2366 | No | ||
| 60 | ACCN4 | 1437091_at | 9628 | 0.134 | 0.2276 | No | ||
| 61 | ZHX2 | 1455210_at | 9967 | 0.104 | 0.2132 | No | ||
| 62 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 10123 | 0.092 | 0.2071 | No | ||
| 63 | HOXD13 | 1422239_at 1440626_at | 10396 | 0.069 | 0.1953 | No | ||
| 64 | CAPZA3 | 1418556_at | 10770 | 0.038 | 0.1786 | No | ||
| 65 | INSRR | 1420564_at | 11054 | 0.012 | 0.1658 | No | ||
| 66 | LHX5 | 1420348_at | 11114 | 0.007 | 0.1631 | No | ||
| 67 | FOXP1 | 1421140_a_at 1421141_a_at 1421142_s_at 1435222_at 1438802_at 1443258_at 1446280_at 1447209_at 1455242_at 1456851_at | 11199 | -0.000 | 0.1593 | No | ||
| 68 | CDKN2C | 1416868_at 1439164_at | 11228 | -0.002 | 0.1580 | No | ||
| 69 | MAPK6 | 1419168_at 1419169_at 1429963_at 1459146_at | 11553 | -0.033 | 0.1435 | No | ||
| 70 | PLRG1 | 1448282_at | 11567 | -0.034 | 0.1433 | No | ||
| 71 | HOXC4 | 1422870_at | 11570 | -0.035 | 0.1435 | No | ||
| 72 | SDF2L1 | 1418206_at | 11675 | -0.044 | 0.1393 | No | ||
| 73 | KEL | 1449057_at | 11879 | -0.061 | 0.1306 | No | ||
| 74 | STARD8 | 1427072_at | 12044 | -0.076 | 0.1239 | No | ||
| 75 | PPM1E | 1434990_at 1445741_at | 12048 | -0.076 | 0.1245 | No | ||
| 76 | ESRRG | 1421747_at 1455267_at 1457896_at | 12225 | -0.091 | 0.1174 | No | ||
| 77 | FGF20 | 1421677_at | 12263 | -0.095 | 0.1167 | No | ||
| 78 | ARPC1A | 1416079_a_at | 12302 | -0.098 | 0.1160 | No | ||
| 79 | PITPNM2 | 1419757_at 1430197_a_at | 12414 | -0.110 | 0.1121 | No | ||
| 80 | NEO1 | 1423513_at 1434931_at 1440789_at 1444295_at 1447693_s_at 1447694_x_at 1459485_at | 12700 | -0.135 | 0.1005 | No | ||
| 81 | SRPK2 | 1417134_at 1417135_at 1417136_s_at 1431372_at 1435746_at 1440080_at 1442241_at 1442980_at 1447755_at 1448603_at | 13008 | -0.165 | 0.0881 | No | ||
| 82 | CCT7 | 1415816_at 1415817_s_at | 13014 | -0.165 | 0.0896 | No | ||
| 83 | PHOX2B | 1422232_at 1455907_x_at | 13035 | -0.167 | 0.0905 | No | ||
| 84 | BHLHB3 | 1421099_at | 13303 | -0.190 | 0.0802 | No | ||
| 85 | SCARF2 | 1434740_at 1438203_at 1438406_at | 13323 | -0.192 | 0.0814 | No | ||
| 86 | ACIN1 | 1416568_a_at 1421197_a_at | 13398 | -0.200 | 0.0801 | No | ||
| 87 | KIF13A | 1439532_s_at 1440578_at 1442252_at 1446026_at 1447853_x_at 1451890_at 1455746_at | 13550 | -0.213 | 0.0754 | No | ||
| 88 | AEBP2 | 1434080_at 1435070_at 1437743_at | 13694 | -0.225 | 0.0712 | No | ||
| 89 | JMJD2D | 1440105_at | 13711 | -0.227 | 0.0729 | No | ||
| 90 | CD36 | 1423166_at 1450883_a_at 1450884_at | 13785 | -0.233 | 0.0720 | No | ||
| 91 | NKX2-2 | 1421112_at | 14062 | -0.262 | 0.0621 | No | ||
| 92 | NSD1 | 1420881_at 1435088_at 1440972_at 1456916_at | 14372 | -0.294 | 0.0510 | No | ||
| 93 | BMP5 | 1421282_at 1421283_at 1444470_x_at 1455851_at | 14394 | -0.296 | 0.0532 | No | ||
| 94 | NTRK1 | 1443990_at | 14398 | -0.296 | 0.0562 | No | ||
| 95 | CHRM1 | 1439611_at 1450833_at | 14520 | -0.308 | 0.0539 | No | ||
| 96 | SCHIP1 | 1423025_a_at 1437700_at 1446421_at 1458588_at | 14533 | -0.309 | 0.0566 | No | ||
| 97 | GATA6 | 1425463_at 1425464_at 1443081_at | 14749 | -0.335 | 0.0503 | No | ||
| 98 | GSC | 1421412_at | 15265 | -0.391 | 0.0308 | No | ||
| 99 | SYTL2 | 1421594_a_at 1456593_at | 15292 | -0.394 | 0.0338 | No | ||
| 100 | WNT7A | 1423367_at 1447647_at 1458334_at | 15379 | -0.403 | 0.0341 | No | ||
| 101 | TBR1 | 1416711_at | 15450 | -0.412 | 0.0352 | No | ||
| 102 | GOLGA1 | 1431120_a_at 1432054_at 1435027_at 1446087_at | 15782 | -0.452 | 0.0248 | No | ||
| 103 | TIAM1 | 1418057_at 1444373_at 1453887_a_at | 16453 | -0.538 | -0.0003 | No | ||
| 104 | PCDH17 | 1436920_at 1446821_at 1453070_at | 17123 | -0.642 | -0.0242 | No | ||
| 105 | DLC1 | 1436173_at 1450206_at 1458220_at 1460602_at | 17419 | -0.688 | -0.0304 | No | ||
| 106 | PMCH | 1429361_at | 17423 | -0.688 | -0.0233 | No | ||
| 107 | GPR126 | 1437408_at 1437409_s_at | 17613 | -0.724 | -0.0243 | No | ||
| 108 | SYT4 | 1415844_at 1415845_at | 18004 | -0.800 | -0.0338 | No | ||
| 109 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 18012 | -0.801 | -0.0257 | No | ||
| 110 | THRA | 1426997_at 1443952_at 1454675_at | 18609 | -0.946 | -0.0430 | No | ||
| 111 | FOXP2 | 1422014_at 1438231_at 1438232_at 1440108_at 1441365_at 1458191_at | 18792 | -0.982 | -0.0410 | No | ||
| 112 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 19004 | -1.017 | -0.0400 | No | ||
| 113 | SPON1 | 1424415_s_at 1441226_at 1451342_at | 19019 | -1.020 | -0.0299 | No | ||
| 114 | SDPR | 1416778_at 1416779_at 1443832_s_at | 19342 | -1.092 | -0.0331 | No | ||
| 115 | TNRC15 | 1439837_at 1446275_at 1451856_at 1457472_at | 19344 | -1.093 | -0.0216 | No | ||
| 116 | STAT5B | 1422102_a_at 1422103_a_at | 19654 | -1.181 | -0.0233 | No | ||
| 117 | LRRTM1 | 1437746_at 1452624_at 1455883_a_at | 19715 | -1.203 | -0.0134 | No | ||
| 118 | ESR1 | 1421244_at 1435663_at 1457877_at 1460591_at | 19849 | -1.253 | -0.0063 | No | ||
| 119 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 20101 | -1.368 | -0.0034 | No | ||
| 120 | TSGA10 | 1436045_at 1447283_at | 20162 | -1.392 | 0.0085 | No | ||
| 121 | ASB4 | 1423422_at 1433919_at | 20467 | -1.544 | 0.0109 | No | ||
| 122 | NLGN2 | 1455143_at 1460013_at | 20618 | -1.639 | 0.0213 | No | ||
| 123 | EPHB2 | 1425015_at 1425016_at 1442471_at 1445417_at 1454022_at | 20648 | -1.657 | 0.0374 | No | ||
| 124 | GGA3 | 1438008_at | 21185 | -2.048 | 0.0344 | No |