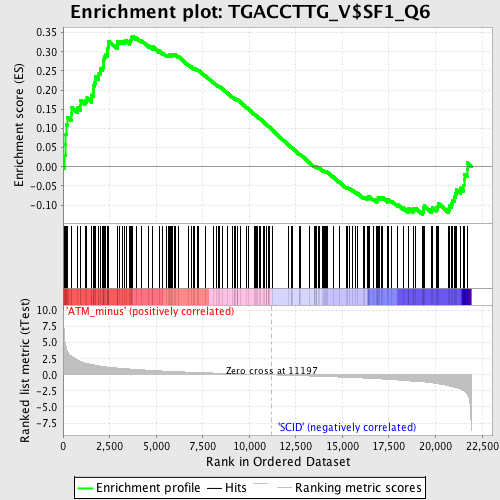
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | TGACCTTG_V$SF1_Q6 |
| Enrichment Score (ES) | 0.3410655 |
| Normalized Enrichment Score (NES) | 1.3520656 |
| Nominal p-value | 0.02852615 |
| FDR q-value | 0.76813793 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AMPD3 | 1422573_at | 82 | 5.554 | 0.0306 | Yes | ||
| 2 | HSPD1 | 1426351_at | 118 | 4.752 | 0.0584 | Yes | ||
| 3 | TNNI1 | 1450813_a_at | 147 | 4.413 | 0.0844 | Yes | ||
| 4 | NDFIP1 | 1424820_a_at 1424821_at 1451493_at | 160 | 4.284 | 0.1104 | Yes | ||
| 5 | NTNG2 | 1432033_at 1450820_a_at | 230 | 3.645 | 0.1298 | Yes | ||
| 6 | GABPA | 1450664_at 1450665_at | 436 | 2.951 | 0.1386 | Yes | ||
| 7 | CACNB2 | 1441521_at 1444693_at 1446632_at 1452476_at 1456401_at | 457 | 2.901 | 0.1556 | Yes | ||
| 8 | UBL3 | 1423461_a_at | 766 | 2.290 | 0.1556 | Yes | ||
| 9 | PNOC | 1425892_a_at | 936 | 2.045 | 0.1605 | Yes | ||
| 10 | SYNGR1 | 1419289_a_at 1434661_at 1453772_at | 940 | 2.041 | 0.1730 | Yes | ||
| 11 | IDH3A | 1422500_at 1422501_s_at 1432016_a_at 1447701_x_at | 1176 | 1.822 | 0.1735 | Yes | ||
| 12 | ZDHHC21 | 1429488_at | 1280 | 1.746 | 0.1796 | Yes | ||
| 13 | MTCH2 | 1430474_a_at 1430969_at 1437806_x_at 1438842_at 1438843_x_at 1448654_at | 1526 | 1.618 | 0.1783 | Yes | ||
| 14 | CS | 1422577_at 1422578_at 1450667_a_at | 1547 | 1.601 | 0.1873 | Yes | ||
| 15 | PHF5A | 1424170_at | 1625 | 1.545 | 0.1933 | Yes | ||
| 16 | VDAC2 | 1415990_at | 1631 | 1.542 | 0.2026 | Yes | ||
| 17 | RNF4 | 1423654_a_at 1445749_at 1451072_a_at | 1638 | 1.539 | 0.2119 | Yes | ||
| 18 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 1671 | 1.526 | 0.2199 | Yes | ||
| 19 | PPP3CC | 1420743_a_at 1430025_at | 1727 | 1.491 | 0.2266 | Yes | ||
| 20 | ATP6AP2 | 1423662_at 1437688_x_at 1439456_x_at | 1747 | 1.481 | 0.2349 | Yes | ||
| 21 | ATP5J | 1416143_at | 1919 | 1.397 | 0.2356 | Yes | ||
| 22 | STK32A | 1456818_at | 1924 | 1.396 | 0.2441 | Yes | ||
| 23 | ATP6V0C | 1416392_a_at 1435732_x_at 1438925_x_at | 1991 | 1.357 | 0.2495 | Yes | ||
| 24 | TIMM9 | 1428010_at 1449886_a_at | 2026 | 1.343 | 0.2562 | Yes | ||
| 25 | KIN | 1427317_at | 2140 | 1.293 | 0.2590 | Yes | ||
| 26 | SDK1 | 1445326_at 1459711_at | 2171 | 1.283 | 0.2656 | Yes | ||
| 27 | ATP1B1 | 1418453_a_at 1423890_x_at 1439036_a_at 1451152_a_at 1455949_at | 2183 | 1.278 | 0.2730 | Yes | ||
| 28 | AK2 | 1448450_at 1448451_at | 2189 | 1.273 | 0.2806 | Yes | ||
| 29 | EEF1A2 | 1418062_at | 2206 | 1.264 | 0.2877 | Yes | ||
| 30 | UCHL1 | 1448260_at | 2282 | 1.231 | 0.2919 | Yes | ||
| 31 | HMGB3 | 1416155_at | 2373 | 1.200 | 0.2952 | Yes | ||
| 32 | TIMM8B | 1431665_a_at | 2393 | 1.194 | 0.3017 | Yes | ||
| 33 | PPP1R16B | 1425414_at 1455080_at | 2404 | 1.188 | 0.3086 | Yes | ||
| 34 | COMTD1 | 1428635_at | 2430 | 1.177 | 0.3147 | Yes | ||
| 35 | SEMA7A | 1422040_at 1459903_at | 2456 | 1.168 | 0.3208 | Yes | ||
| 36 | LAMB2 | 1416513_at | 2460 | 1.168 | 0.3279 | Yes | ||
| 37 | ARF3 | 1421789_s_at 1423973_a_at 1434787_at 1437331_a_at 1456007_at | 2902 | 1.046 | 0.3141 | Yes | ||
| 38 | DNER | 1423671_at 1456379_x_at | 2906 | 1.045 | 0.3204 | Yes | ||
| 39 | RANBP1 | 1422547_at | 2915 | 1.042 | 0.3265 | Yes | ||
| 40 | ZRANB1 | 1415712_at 1427200_at 1436228_at | 3020 | 1.019 | 0.3280 | Yes | ||
| 41 | PCDH7 | 1427592_at 1437442_at 1449249_at 1456214_at | 3165 | 0.994 | 0.3276 | Yes | ||
| 42 | FZD9 | 1427529_at | 3278 | 0.968 | 0.3284 | Yes | ||
| 43 | TCF2 | 1421224_a_at 1441484_at 1451687_a_at | 3393 | 0.945 | 0.3290 | Yes | ||
| 44 | ATP5G1 | 1416020_a_at 1444874_at | 3587 | 0.902 | 0.3257 | Yes | ||
| 45 | PHACTR3 | 1429651_at | 3609 | 0.898 | 0.3303 | Yes | ||
| 46 | SLC30A3 | 1460654_at | 3649 | 0.888 | 0.3340 | Yes | ||
| 47 | PRPSAP1 | 1428182_at | 3666 | 0.884 | 0.3388 | Yes | ||
| 48 | SH3GL2 | 1418791_at 1418792_at 1449228_at | 3734 | 0.870 | 0.3411 | Yes | ||
| 49 | MTX2 | 1430500_s_at | 3918 | 0.835 | 0.3378 | No | ||
| 50 | PDHX | 1456090_at 1459434_at | 4219 | 0.781 | 0.3289 | No | ||
| 51 | FSTL3 | 1422803_at | 4601 | 0.706 | 0.3157 | No | ||
| 52 | VAMP2 | 1420833_at 1420834_at | 4806 | 0.673 | 0.3105 | No | ||
| 53 | RHOT1 | 1423533_a_at 1440853_at | 4809 | 0.673 | 0.3146 | No | ||
| 54 | ATP5G3 | 1454661_at | 5155 | 0.614 | 0.3025 | No | ||
| 55 | RPH3A | 1434635_at 1449961_at | 5363 | 0.585 | 0.2966 | No | ||
| 56 | RILP | 1457696_at | 5548 | 0.558 | 0.2916 | No | ||
| 57 | CCNE1 | 1416492_at 1441910_x_at | 5660 | 0.545 | 0.2899 | No | ||
| 58 | EPN3 | 1424960_at | 5689 | 0.541 | 0.2920 | No | ||
| 59 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 5790 | 0.528 | 0.2906 | No | ||
| 60 | MYL3 | 1427768_s_at 1427769_x_at 1428266_at | 5846 | 0.519 | 0.2913 | No | ||
| 61 | NPTX2 | 1420720_at 1449960_at | 5855 | 0.518 | 0.2942 | No | ||
| 62 | SNPH | 1435210_s_at | 5959 | 0.506 | 0.2926 | No | ||
| 63 | MRPL47 | 1429054_at 1443915_at 1453927_at | 6008 | 0.501 | 0.2935 | No | ||
| 64 | SLC37A2 | 1452492_a_at | 6191 | 0.479 | 0.2881 | No | ||
| 65 | HTF9C | 1434244_x_at 1441920_x_at 1448114_a_at 1448115_at 1453995_a_at 1457620_at | 6734 | 0.416 | 0.2657 | No | ||
| 66 | ATP5C1 | 1416058_s_at 1438809_at | 6876 | 0.400 | 0.2617 | No | ||
| 67 | CDH16 | 1448906_at | 7026 | 0.383 | 0.2572 | No | ||
| 68 | APP | 1420621_a_at 1427442_a_at 1438373_at 1438374_x_at 1444705_at 1447347_at 1458525_at | 7070 | 0.379 | 0.2576 | No | ||
| 69 | CNTN2 | 1435165_at 1435166_at 1450523_at 1456962_at | 7227 | 0.360 | 0.2527 | No | ||
| 70 | LRFN5 | 1441502_at 1454687_at | 7288 | 0.355 | 0.2521 | No | ||
| 71 | MGST3 | 1448300_at 1450563_at | 7655 | 0.318 | 0.2373 | No | ||
| 72 | CA2 | 1448752_at | 8095 | 0.277 | 0.2188 | No | ||
| 73 | INSR | 1421380_at 1450225_at | 8247 | 0.262 | 0.2135 | No | ||
| 74 | WDFY3 | 1427456_at 1428517_at 1445718_at 1446109_at 1447433_at 1458952_at 1459287_at 1459414_at | 8352 | 0.250 | 0.2102 | No | ||
| 75 | AUH | 1420776_a_at 1430487_at 1431124_at 1444433_at 1445496_at 1445800_at 1447952_at 1458436_at | 8424 | 0.242 | 0.2085 | No | ||
| 76 | ASTN2 | 1442857_at 1443401_at 1450309_at | 8548 | 0.230 | 0.2043 | No | ||
| 77 | PHF15 | 1430081_at 1455345_at | 8828 | 0.205 | 0.1927 | No | ||
| 78 | ELAVL3 | 1452524_a_at | 9091 | 0.182 | 0.1818 | No | ||
| 79 | VSNL1 | 1420955_at 1450055_at | 9222 | 0.170 | 0.1769 | No | ||
| 80 | RIN2 | 1426368_at 1441209_at 1442829_at | 9227 | 0.170 | 0.1777 | No | ||
| 81 | ATP6V1B1 | 1419373_at | 9268 | 0.166 | 0.1769 | No | ||
| 82 | KIRREL3 | 1431402_at 1441357_at 1452728_at | 9349 | 0.158 | 0.1742 | No | ||
| 83 | MEF2D | 1421388_at 1434487_at 1436642_x_at 1437300_at 1458901_at | 9351 | 0.157 | 0.1751 | No | ||
| 84 | CHGA | 1418149_at | 9356 | 0.157 | 0.1759 | No | ||
| 85 | ZNF289 | 1439460_a_at 1448672_a_at | 9521 | 0.143 | 0.1693 | No | ||
| 86 | CAMK2G | 1423941_at 1423942_a_at | 9873 | 0.112 | 0.1538 | No | ||
| 87 | HOXC10 | 1439798_at | 9977 | 0.103 | 0.1498 | No | ||
| 88 | DIRAS1 | 1425748_at 1451820_at 1455526_at | 10257 | 0.081 | 0.1374 | No | ||
| 89 | RHCG | 1417362_at | 10331 | 0.074 | 0.1345 | No | ||
| 90 | CHAT | 1440070_at | 10372 | 0.070 | 0.1331 | No | ||
| 91 | SCN5A | 1422194_at 1458813_at | 10444 | 0.064 | 0.1303 | No | ||
| 92 | AFG3L2 | 1427206_at 1427207_s_at 1454003_at | 10553 | 0.056 | 0.1257 | No | ||
| 93 | KCNK1 | 1448690_at 1455896_a_at | 10598 | 0.052 | 0.1240 | No | ||
| 94 | TFRC | 1422966_a_at 1422967_a_at 1452661_at AFFX-TransRecMur/X57349_3_at AFFX-TransRecMur/X57349_5_at AFFX-TransRecMur/X57349_M_at | 10759 | 0.040 | 0.1169 | No | ||
| 95 | CASQ2 | 1422529_s_at | 10839 | 0.032 | 0.1134 | No | ||
| 96 | GABRA3 | 1421263_at | 10908 | 0.027 | 0.1105 | No | ||
| 97 | FGF9 | 1420795_at 1438718_at 1445258_at | 10930 | 0.025 | 0.1097 | No | ||
| 98 | EPS8L2 | 1417843_s_at | 11023 | 0.014 | 0.1055 | No | ||
| 99 | RAB3A | 1422589_at 1459980_x_at | 11036 | 0.013 | 0.1050 | No | ||
| 100 | SLC38A3 | 1418706_at 1446013_at | 11067 | 0.011 | 0.1037 | No | ||
| 101 | NXPH4 | 1429753_at | 11269 | -0.007 | 0.0945 | No | ||
| 102 | SV2A | 1423406_at | 12078 | -0.079 | 0.0579 | No | ||
| 103 | SEMA3F | 1420508_at 1425840_a_at 1438947_x_at | 12272 | -0.096 | 0.0496 | No | ||
| 104 | FNDC5 | 1435115_at 1453135_at | 12275 | -0.096 | 0.0501 | No | ||
| 105 | CRSP7 | 1452282_at 1458377_at | 12338 | -0.102 | 0.0479 | No | ||
| 106 | NRXN2 | 1435907_at 1435908_at 1455499_at | 12676 | -0.132 | 0.0332 | No | ||
| 107 | NCDN | 1416852_a_at 1416853_at | 12697 | -0.134 | 0.0331 | No | ||
| 108 | HCN1 | 1450193_at | 12754 | -0.140 | 0.0314 | No | ||
| 109 | KIF5A | 1434670_at | 13246 | -0.186 | 0.0100 | No | ||
| 110 | COX5B | 1416902_a_at 1435613_x_at 1436149_at 1448514_at 1454716_x_at 1456588_x_at | 13475 | -0.205 | 0.0008 | No | ||
| 111 | DSCR1L1 | 1421425_a_at 1450243_a_at | 13500 | -0.208 | 0.0010 | No | ||
| 112 | CYC1 | 1416604_at | 13567 | -0.214 | -0.0007 | No | ||
| 113 | MNT | 1418192_at | 13597 | -0.216 | -0.0007 | No | ||
| 114 | MAEA | 1438903_at | 13601 | -0.217 | 0.0005 | No | ||
| 115 | NDUFB5 | 1417102_a_at 1448589_at | 13689 | -0.224 | -0.0021 | No | ||
| 116 | RNF139 | 1429425_at 1429426_at | 13766 | -0.231 | -0.0042 | No | ||
| 117 | POLR3D | 1424565_at 1424566_s_at | 13932 | -0.249 | -0.0102 | No | ||
| 118 | DNAJC11 | 1433880_at 1433881_at 1439763_at 1446686_at 1447055_at | 13967 | -0.253 | -0.0102 | No | ||
| 119 | ATP5B | 1416829_at | 14028 | -0.259 | -0.0114 | No | ||
| 120 | SLITRK1 | 1428089_at | 14100 | -0.267 | -0.0130 | No | ||
| 121 | CATSPER2 | 1425309_at 1445264_at | 14132 | -0.270 | -0.0127 | No | ||
| 122 | CEL | 1417257_at | 14199 | -0.277 | -0.0141 | No | ||
| 123 | KCNG4 | 1428536_at | 14512 | -0.307 | -0.0265 | No | ||
| 124 | STAC2 | 1424581_at | 14840 | -0.345 | -0.0394 | No | ||
| 125 | FANCD2 | 1430916_at 1438339_at 1439091_at 1442092_at | 15206 | -0.385 | -0.0538 | No | ||
| 126 | TUBB4 | 1423221_at | 15248 | -0.390 | -0.0533 | No | ||
| 127 | MAL | 1417275_at 1432558_a_at | 15378 | -0.403 | -0.0567 | No | ||
| 128 | NNAT | 1423506_a_at | 15539 | -0.423 | -0.0614 | No | ||
| 129 | UQCRC1 | 1428782_a_at | 15710 | -0.444 | -0.0665 | No | ||
| 130 | SLC6A8 | 1417116_at 1448596_at | 15826 | -0.457 | -0.0689 | No | ||
| 131 | YWHAH | 1416004_at | 16143 | -0.498 | -0.0804 | No | ||
| 132 | TBX6 | 1449868_at | 16207 | -0.505 | -0.0802 | No | ||
| 133 | TMEM28 | 1435138_at 1443271_at 1445869_at 1446912_at 1459483_at | 16333 | -0.523 | -0.0827 | No | ||
| 134 | MASP1 | 1419677_at 1425985_s_at 1438602_s_at 1438603_x_at 1455346_at | 16375 | -0.527 | -0.0813 | No | ||
| 135 | BLCAP | 1420631_a_at | 16382 | -0.528 | -0.0783 | No | ||
| 136 | ADCY1 | 1445112_at 1445359_at 1456487_at 1457251_x_at 1457329_at | 16429 | -0.534 | -0.0771 | No | ||
| 137 | HOXC13 | 1425874_at | 16679 | -0.574 | -0.0850 | No | ||
| 138 | IBRDC3 | 1432478_a_at 1435226_at | 16845 | -0.597 | -0.0889 | No | ||
| 139 | SNTG1 | 1432287_a_at 1440598_at | 16890 | -0.602 | -0.0872 | No | ||
| 140 | KCNMB2 | 1426322_a_at 1431844_at | 16909 | -0.605 | -0.0843 | No | ||
| 141 | ABHD3 | 1417946_at | 16915 | -0.606 | -0.0807 | No | ||
| 142 | GBAS | 1419484_a_at 1444503_at 1446137_at | 17007 | -0.621 | -0.0811 | No | ||
| 143 | RGS7 | 1450659_at | 17089 | -0.634 | -0.0809 | No | ||
| 144 | CKB | 1455106_a_at | 17140 | -0.645 | -0.0792 | No | ||
| 145 | NTF3 | 1434802_s_at 1450803_at | 17431 | -0.691 | -0.0882 | No | ||
| 146 | SLC2A4 | 1415958_at 1415959_at | 17460 | -0.695 | -0.0852 | No | ||
| 147 | COX4I1 | 1448322_a_at | 17661 | -0.733 | -0.0899 | No | ||
| 148 | VASP | 1451097_at | 17964 | -0.792 | -0.0989 | No | ||
| 149 | MSI2 | 1421230_a_at 1421231_at 1435520_at 1435521_at 1436818_a_at 1441185_at 1441373_at 1443008_at 1443530_at 1446022_at 1458136_at 1458542_at | 18273 | -0.866 | -0.1077 | No | ||
| 150 | PANK1 | 1418715_at 1429813_at 1429814_at 1431028_a_at 1457110_at | 18528 | -0.927 | -0.1136 | No | ||
| 151 | SDHD | 1428235_at 1437489_x_at | 18565 | -0.936 | -0.1095 | No | ||
| 152 | PCTK1 | 1415956_a_at 1438314_at 1443296_at 1460169_a_at | 18793 | -0.982 | -0.1138 | No | ||
| 153 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 18835 | -0.989 | -0.1096 | No | ||
| 154 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 18943 | -1.008 | -0.1083 | No | ||
| 155 | WWP1 | 1427097_at 1427098_at 1446364_at 1446853_at 1452299_at | 19302 | -1.078 | -0.1180 | No | ||
| 156 | CTSD | 1448118_a_at | 19337 | -1.091 | -0.1128 | No | ||
| 157 | TRAP1 | 1416170_at 1437379_x_at | 19356 | -1.097 | -0.1069 | No | ||
| 158 | IMMT | 1429533_at 1429534_a_at 1433470_a_at 1437126_at | 19395 | -1.104 | -0.1018 | No | ||
| 159 | AMY2A | 1457367_at | 19794 | -1.231 | -0.1125 | No | ||
| 160 | ABTB2 | 1433453_a_at 1433454_at 1443497_at 1444771_at | 19830 | -1.244 | -0.1064 | No | ||
| 161 | ANKRD28 | 1434630_at 1454801_at | 20035 | -1.334 | -0.1075 | No | ||
| 162 | RBMS1 | 1418703_at 1434005_at 1441437_at 1458578_at 1459968_at | 20132 | -1.382 | -0.1034 | No | ||
| 163 | FBXO18 | 1452153_at | 20152 | -1.388 | -0.0957 | No | ||
| 164 | RBMX | 1416177_at 1416354_at 1416355_at 1426863_at 1437847_x_at | 20699 | -1.696 | -0.1103 | No | ||
| 165 | RAPGEF5 | 1445255_at 1455137_at 1455840_at | 20761 | -1.732 | -0.1023 | No | ||
| 166 | RANBP10 | 1424584_a_at 1424585_at 1446446_at | 20847 | -1.794 | -0.0952 | No | ||
| 167 | UBE1 | 1442767_s_at 1448116_at | 20931 | -1.864 | -0.0874 | No | ||
| 168 | DEXI | 1460174_at | 20999 | -1.919 | -0.0786 | No | ||
| 169 | CHD2 | 1444246_at 1445843_at | 21056 | -1.957 | -0.0691 | No | ||
| 170 | ESRRA | 1442864_at 1460652_at | 21130 | -2.017 | -0.0600 | No | ||
| 171 | GABARAPL1 | 1416418_at 1416419_s_at 1442572_at | 21332 | -2.220 | -0.0555 | No | ||
| 172 | HSPE1 | 1450668_s_at | 21520 | -2.483 | -0.0487 | No | ||
| 173 | TOMM40 | 1426118_a_at 1459680_at | 21532 | -2.494 | -0.0338 | No | ||
| 174 | ACO2 | 1436934_s_at 1442102_at 1451002_at | 21552 | -2.526 | -0.0190 | No | ||
| 175 | MADD | 1425735_at 1455502_at | 21698 | -2.944 | -0.0075 | No | ||
| 176 | TBC1D15 | 1416060_at 1416061_at 1416062_at | 21705 | -2.967 | 0.0106 | No |

