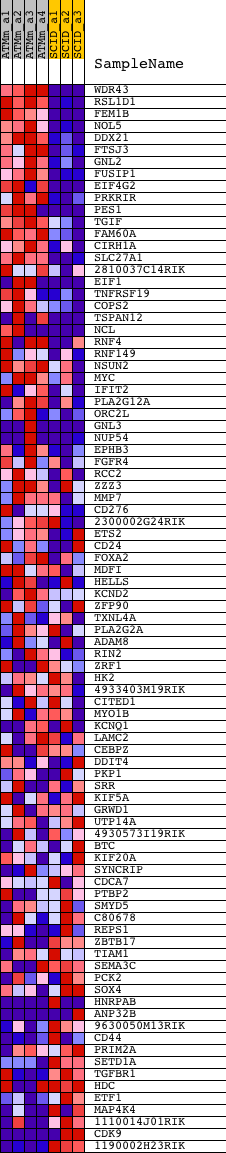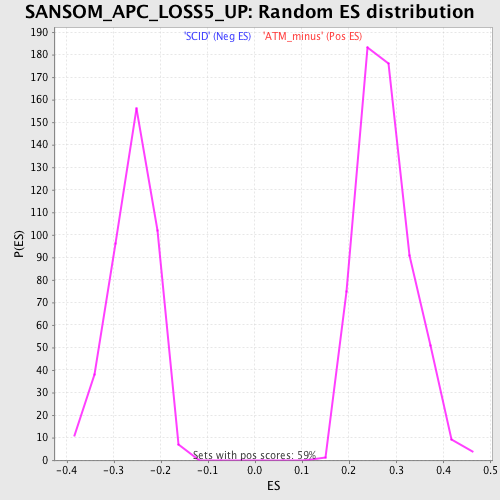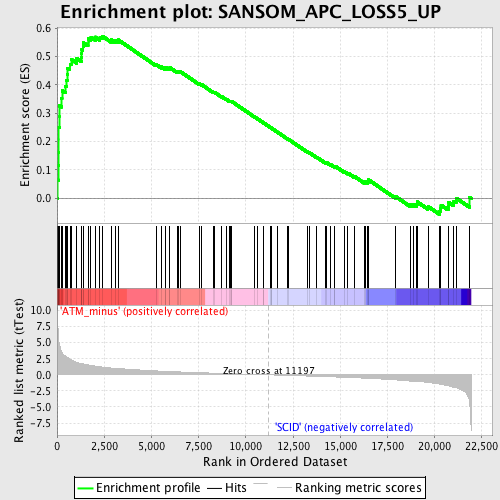
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | SANSOM_APC_LOSS5_UP |
| Enrichment Score (ES) | 0.5724142 |
| Normalized Enrichment Score (NES) | 2.0744605 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.021465454 |
| FWER p-Value | 0.018 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WDR43 | 1428389_s_at 1428390_at | 33 | 8.025 | 0.0653 | Yes | ||
| 2 | RSL1D1 | 1456383_at | 61 | 6.230 | 0.1159 | Yes | ||
| 3 | FEM1B | 1418323_at 1418324_at | 81 | 5.632 | 0.1620 | Yes | ||
| 4 | NOL5 | 1450986_at | 90 | 5.391 | 0.2065 | Yes | ||
| 5 | DDX21 | 1448270_at 1448271_a_at | 91 | 5.390 | 0.2514 | Yes | ||
| 6 | FTSJ3 | 1451026_at | 127 | 4.654 | 0.2885 | Yes | ||
| 7 | GNL2 | 1426579_at | 134 | 4.575 | 0.3263 | Yes | ||
| 8 | FUSIP1 | 1418526_at 1418527_a_at 1423982_at 1449121_at | 229 | 3.646 | 0.3524 | Yes | ||
| 9 | EIF4G2 | 1415863_at 1452758_s_at | 260 | 3.482 | 0.3800 | Yes | ||
| 10 | PRKRIR | 1426482_at 1426483_at | 441 | 2.939 | 0.3962 | Yes | ||
| 11 | PES1 | 1416172_at 1416173_at | 507 | 2.800 | 0.4166 | Yes | ||
| 12 | TGIF | 1422286_a_at | 532 | 2.734 | 0.4383 | Yes | ||
| 13 | FAM60A | 1448126_at | 565 | 2.676 | 0.4591 | Yes | ||
| 14 | CIRH1A | 1423884_at | 682 | 2.420 | 0.4739 | Yes | ||
| 15 | SLC27A1 | 1422811_at 1435658_at | 755 | 2.305 | 0.4898 | Yes | ||
| 16 | 2810037C14RIK | 1417062_at | 1038 | 1.942 | 0.4931 | Yes | ||
| 17 | EIF1 | 1423798_a_at 1423799_at 1424343_a_at 1424344_s_at | 1279 | 1.747 | 0.4966 | Yes | ||
| 18 | TNFRSF19 | 1415921_a_at 1425212_a_at 1427600_at 1445949_at 1448147_at | 1295 | 1.739 | 0.5104 | Yes | ||
| 19 | COPS2 | 1423459_at 1437039_at | 1310 | 1.731 | 0.5242 | Yes | ||
| 20 | TSPAN12 | 1438050_x_at 1454604_s_at | 1371 | 1.699 | 0.5356 | Yes | ||
| 21 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 1383 | 1.694 | 0.5492 | Yes | ||
| 22 | RNF4 | 1423654_a_at 1445749_at 1451072_a_at | 1638 | 1.539 | 0.5504 | Yes | ||
| 23 | RNF149 | 1429321_at | 1640 | 1.538 | 0.5632 | Yes | ||
| 24 | NSUN2 | 1423850_at 1453759_at | 1791 | 1.459 | 0.5684 | Yes | ||
| 25 | MYC | 1424942_a_at | 2017 | 1.346 | 0.5693 | Yes | ||
| 26 | IFIT2 | 1418293_at | 2270 | 1.236 | 0.5681 | Yes | ||
| 27 | PLA2G12A | 1452026_a_at | 2394 | 1.194 | 0.5724 | Yes | ||
| 28 | ORC2L | 1418225_at 1418226_at 1418227_at | 2868 | 1.056 | 0.5596 | No | ||
| 29 | GNL3 | 1433656_a_at 1440766_at | 3077 | 1.008 | 0.5584 | No | ||
| 30 | NUP54 | 1432539_a_at 1433580_at 1438951_x_at 1447253_x_at 1459651_s_at | 3241 | 0.977 | 0.5591 | No | ||
| 31 | EPHB3 | 1451550_at | 5244 | 0.602 | 0.4725 | No | ||
| 32 | FGFR4 | 1427776_a_at 1427777_x_at 1427845_at 1427846_x_at | 5529 | 0.561 | 0.4641 | No | ||
| 33 | RCC2 | 1426897_at 1452226_at | 5734 | 0.535 | 0.4593 | No | ||
| 34 | ZZZ3 | 1434332_at 1438487_s_at 1441748_at | 5753 | 0.533 | 0.4629 | No | ||
| 35 | MMP7 | 1449478_at | 5961 | 0.506 | 0.4576 | No | ||
| 36 | CD276 | 1417599_at | 5973 | 0.505 | 0.4613 | No | ||
| 37 | 2300002G24RIK | 1453092_at | 6379 | 0.454 | 0.4466 | No | ||
| 38 | ETS2 | 1416268_at 1437809_x_at 1447685_x_at | 6434 | 0.449 | 0.4478 | No | ||
| 39 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 6541 | 0.437 | 0.4466 | No | ||
| 40 | FOXA2 | 1422833_at | 7521 | 0.330 | 0.4045 | No | ||
| 41 | MDFI | 1420713_a_at 1425924_at | 7643 | 0.319 | 0.4017 | No | ||
| 42 | HELLS | 1417541_at 1430139_at 1453361_at | 8290 | 0.257 | 0.3742 | No | ||
| 43 | KCND2 | 1422834_at 1422835_at 1446065_at 1447764_at 1450773_at 1459288_at | 8317 | 0.254 | 0.3752 | No | ||
| 44 | ZFP90 | 1440334_at 1449126_at | 8709 | 0.215 | 0.3591 | No | ||
| 45 | TXNL4A | 1419179_at | 8958 | 0.193 | 0.3493 | No | ||
| 46 | PLA2G2A | 1450128_at | 9128 | 0.179 | 0.3431 | No | ||
| 47 | ADAM8 | 1416871_at | 9178 | 0.175 | 0.3423 | No | ||
| 48 | RIN2 | 1426368_at 1441209_at 1442829_at | 9227 | 0.170 | 0.3415 | No | ||
| 49 | ZRF1 | 1417657_s_at 1425910_at 1448794_s_at 1454219_at | 9261 | 0.166 | 0.3414 | No | ||
| 50 | HK2 | 1422612_at | 10461 | 0.063 | 0.2870 | No | ||
| 51 | 4933403M19RIK | 1429999_at | 10622 | 0.050 | 0.2801 | No | ||
| 52 | CITED1 | 1449031_at | 10935 | 0.024 | 0.2660 | No | ||
| 53 | MYO1B | 1427450_x_at 1441210_at 1447364_x_at 1448989_a_at 1448990_a_at 1458007_at 1459679_s_at | 10947 | 0.023 | 0.2657 | No | ||
| 54 | KCNQ1 | 1441588_at 1442029_at 1446255_at 1447418_at 1449464_at 1457781_at 1459443_at | 11291 | -0.009 | 0.2501 | No | ||
| 55 | LAMC2 | 1421279_at 1452505_at | 11330 | -0.013 | 0.2485 | No | ||
| 56 | CEBPZ | 1420497_a_at 1429962_at | 11369 | -0.016 | 0.2469 | No | ||
| 57 | DDIT4 | 1428306_at | 11672 | -0.044 | 0.2334 | No | ||
| 58 | PKP1 | 1449586_at | 12221 | -0.091 | 0.2091 | No | ||
| 59 | SRR | 1419664_at 1438549_a_at 1438550_x_at 1453421_at 1455045_at 1458393_at | 12279 | -0.096 | 0.2073 | No | ||
| 60 | KIF5A | 1434670_at | 13246 | -0.186 | 0.1646 | No | ||
| 61 | GRWD1 | 1436887_x_at 1452171_at 1455841_s_at | 13371 | -0.197 | 0.1606 | No | ||
| 62 | UTP14A | 1432250_at 1435099_at | 13736 | -0.228 | 0.1458 | No | ||
| 63 | 4930573I19RIK | 1439502_at | 14213 | -0.278 | 0.1264 | No | ||
| 64 | BTC | 1421161_at 1435541_at | 14250 | -0.282 | 0.1271 | No | ||
| 65 | KIF20A | 1449207_a_at | 14487 | -0.305 | 0.1188 | No | ||
| 66 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 14703 | -0.329 | 0.1117 | No | ||
| 67 | CDCA7 | 1428069_at 1445681_at | 14713 | -0.330 | 0.1140 | No | ||
| 68 | PTBP2 | 1423470_at 1423471_at 1444746_at 1446490_at 1458757_at | 15201 | -0.385 | 0.0949 | No | ||
| 69 | SMYD5 | 1425479_at 1447749_at 1451722_s_at | 15403 | -0.406 | 0.0891 | No | ||
| 70 | C80678 | 1419953_at 1449680_at | 15764 | -0.450 | 0.0764 | No | ||
| 71 | REPS1 | 1416216_at 1440822_x_at 1446001_at | 16269 | -0.514 | 0.0576 | No | ||
| 72 | ZBTB17 | 1416224_at 1443041_at | 16308 | -0.519 | 0.0602 | No | ||
| 73 | TIAM1 | 1418057_at 1444373_at 1453887_a_at | 16453 | -0.538 | 0.0581 | No | ||
| 74 | SEMA3C | 1420696_at 1429348_at | 16473 | -0.542 | 0.0617 | No | ||
| 75 | PCK2 | 1425615_a_at 1452620_at | 16499 | -0.544 | 0.0651 | No | ||
| 76 | SOX4 | 1455867_at 1458332_x_at | 17932 | -0.787 | 0.0061 | No | ||
| 77 | HNRPAB | 1415914_at 1426114_at 1431349_at 1448144_at 1453849_s_at 1455855_x_at 1459880_at | 18694 | -0.964 | -0.0207 | No | ||
| 78 | ANP32B | 1417082_at 1446091_at | 18871 | -0.997 | -0.0204 | No | ||
| 79 | 9630050M13RIK | 1438402_at | 19058 | -1.026 | -0.0204 | No | ||
| 80 | CD44 | 1423760_at 1434376_at 1443265_at 1452483_a_at | 19073 | -1.028 | -0.0125 | No | ||
| 81 | PRIM2A | 1418035_a_at 1418036_at 1445474_at | 19643 | -1.178 | -0.0287 | No | ||
| 82 | SETD1A | 1427116_at | 20262 | -1.439 | -0.0450 | No | ||
| 83 | TGFBR1 | 1420893_a_at 1420894_at 1420895_at 1446946_at | 20289 | -1.457 | -0.0341 | No | ||
| 84 | HDC | 1451796_s_at 1454713_s_at | 20331 | -1.477 | -0.0237 | No | ||
| 85 | ETF1 | 1420023_at 1420024_s_at 1424013_at 1440629_at 1451208_at | 20713 | -1.702 | -0.0269 | No | ||
| 86 | MAP4K4 | 1422615_at 1440609_at 1448050_s_at 1459912_at | 20726 | -1.711 | -0.0132 | No | ||
| 87 | 1110014J01RIK | 1444383_at 1448141_at 1456814_at | 21013 | -1.931 | -0.0102 | No | ||
| 88 | CDK9 | 1417269_at 1457240_at | 21134 | -2.017 | 0.0011 | No | ||
| 89 | 1190002H23RIK | 1418003_at 1438511_a_at | 21843 | -4.276 | 0.0043 | No |