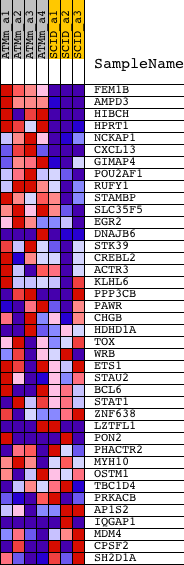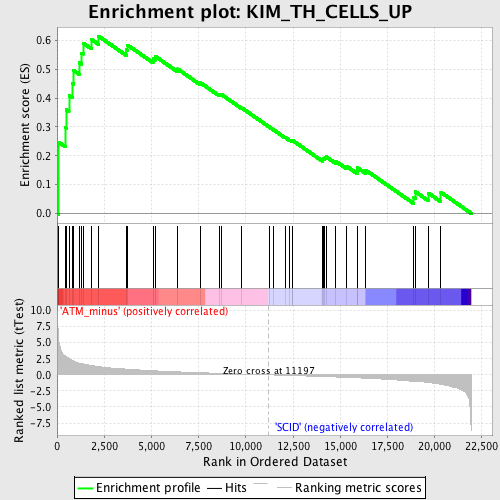
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | KIM_TH_CELLS_UP |
| Enrichment Score (ES) | 0.61503416 |
| Normalized Enrichment Score (NES) | 1.8960778 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09142215 |
| FWER p-Value | 0.37 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FEM1B | 1418323_at 1418324_at | 81 | 5.632 | 0.1227 | Yes | ||
| 2 | AMPD3 | 1422573_at | 82 | 5.554 | 0.2474 | Yes | ||
| 3 | HIBCH | 1451511_at 1451512_s_at | 429 | 2.962 | 0.2982 | Yes | ||
| 4 | HPRT1 | 1448736_a_at | 478 | 2.850 | 0.3599 | Yes | ||
| 5 | NCKAP1 | 1452196_a_at | 630 | 2.512 | 0.4094 | Yes | ||
| 6 | CXCL13 | 1417851_at 1448859_at | 801 | 2.244 | 0.4521 | Yes | ||
| 7 | GIMAP4 | 1424374_at 1424375_s_at | 882 | 2.113 | 0.4958 | Yes | ||
| 8 | POU2AF1 | 1416957_at | 1188 | 1.811 | 0.5226 | Yes | ||
| 9 | RUFY1 | 1434131_at 1447391_at 1457340_at | 1303 | 1.734 | 0.5563 | Yes | ||
| 10 | STAMBP | 1416317_a_at 1431962_a_at | 1378 | 1.697 | 0.5910 | Yes | ||
| 11 | SLC35F5 | 1452059_at | 1824 | 1.446 | 0.6032 | Yes | ||
| 12 | EGR2 | 1427682_a_at 1427683_at | 2191 | 1.273 | 0.6150 | Yes | ||
| 13 | DNAJB6 | 1429776_a_at 1429777_at 1434035_at 1435701_at 1442217_at 1448234_at | 3668 | 0.884 | 0.5675 | No | ||
| 14 | STK39 | 1419550_a_at 1419551_s_at 1431434_at 1445431_at | 3733 | 0.870 | 0.5841 | No | ||
| 15 | CREBL2 | 1435085_at 1442738_at | 5083 | 0.627 | 0.5366 | No | ||
| 16 | ACTR3 | 1426392_a_at 1434968_a_at 1452051_at | 5186 | 0.609 | 0.5456 | No | ||
| 17 | KLHL6 | 1437886_at | 6385 | 0.454 | 0.5011 | No | ||
| 18 | PPP3CB | 1427468_at 1428473_at 1428474_at 1433835_at 1446149_at 1459378_at | 7597 | 0.322 | 0.4530 | No | ||
| 19 | PAWR | 1444808_at 1445390_at | 8581 | 0.228 | 0.4132 | No | ||
| 20 | CHGB | 1415885_at | 8680 | 0.218 | 0.4136 | No | ||
| 21 | HDHD1A | 1420775_at | 9788 | 0.119 | 0.3657 | No | ||
| 22 | TOX | 1425483_at 1425484_at 1442039_at 1446950_at 1458974_at | 11260 | -0.005 | 0.2987 | No | ||
| 23 | WRB | 1430312_at 1460446_at | 11468 | -0.025 | 0.2898 | No | ||
| 24 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 12075 | -0.079 | 0.2639 | No | ||
| 25 | STAU2 | 1425533_a_at 1425534_at 1450916_at | 12330 | -0.101 | 0.2545 | No | ||
| 26 | BCL6 | 1421818_at 1450381_a_at | 12464 | -0.113 | 0.2510 | No | ||
| 27 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 12475 | -0.115 | 0.2531 | No | ||
| 28 | ZNF638 | 1417791_a_at 1417792_at 1447983_at 1458523_at | 14045 | -0.261 | 0.1873 | No | ||
| 29 | LZTFL1 | 1417170_at 1428974_s_at 1431323_at 1435514_at 1440432_at 1460200_s_at | 14089 | -0.265 | 0.1913 | No | ||
| 30 | PON2 | 1425791_at 1429019_s_at 1429020_at 1450686_at | 14187 | -0.276 | 0.1931 | No | ||
| 31 | PHACTR2 | 1452404_at | 14243 | -0.281 | 0.1969 | No | ||
| 32 | MYH10 | 1441057_at 1452740_at | 14739 | -0.333 | 0.1818 | No | ||
| 33 | OSTM1 | 1428334_at 1438521_at | 15317 | -0.396 | 0.1643 | No | ||
| 34 | TBC1D4 | 1435292_at 1455903_at | 15904 | -0.466 | 0.1480 | No | ||
| 35 | PRKACB | 1420610_at 1420611_at 1457197_at | 15911 | -0.466 | 0.1582 | No | ||
| 36 | AP1S2 | 1437998_at 1447903_x_at 1452657_at 1456252_x_at 1460036_at | 16341 | -0.524 | 0.1504 | No | ||
| 37 | IQGAP1 | 1417379_at 1417380_at 1431395_a_at 1431396_at 1434998_at 1445724_at 1458124_at | 18896 | -1.001 | 0.0562 | No | ||
| 38 | MDM4 | 1419558_at 1453982_at 1460542_s_at | 18975 | -1.012 | 0.0753 | No | ||
| 39 | CPSF2 | 1420936_s_at 1420937_at 1431089_at 1443083_at | 19667 | -1.186 | 0.0704 | No | ||
| 40 | SH2D1A | 1449393_at | 20329 | -1.477 | 0.0734 | No |