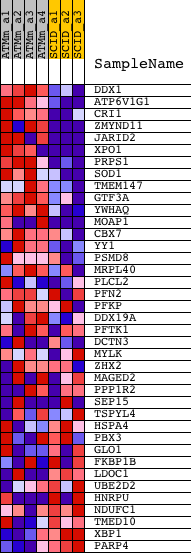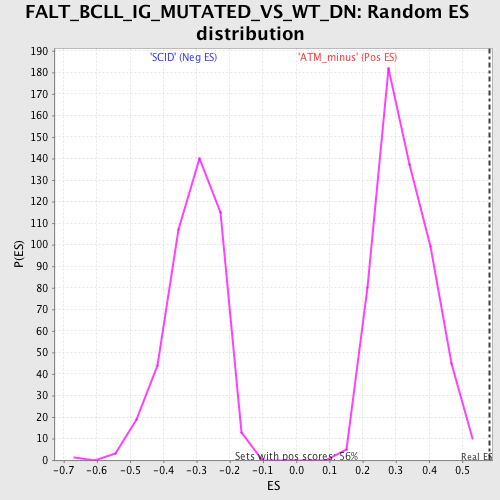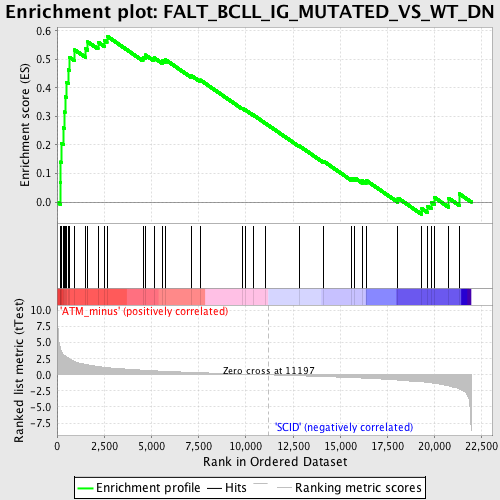
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | FALT_BCLL_IG_MUTATED_VS_WT_DN |
| Enrichment Score (ES) | 0.58112144 |
| Normalized Enrichment Score (NES) | 1.7984828 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.12704417 |
| FWER p-Value | 0.845 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDX1 | 1415915_at 1440816_x_at | 179 | 4.024 | 0.0672 | Yes | ||
| 2 | ATP6V1G1 | 1423255_at | 191 | 3.929 | 0.1404 | Yes | ||
| 3 | CRI1 | 1416614_at 1448405_a_at 1448406_at | 242 | 3.591 | 0.2054 | Yes | ||
| 4 | ZMYND11 | 1426531_at 1426532_at 1436153_a_at 1444461_at 1446182_at | 340 | 3.152 | 0.2600 | Yes | ||
| 5 | JARID2 | 1422698_s_at 1446082_at 1450710_at 1456661_at | 378 | 3.057 | 0.3156 | Yes | ||
| 6 | XPO1 | 1418442_at 1418443_at 1448070_at | 421 | 2.975 | 0.3694 | Yes | ||
| 7 | PRPS1 | 1416052_at | 486 | 2.833 | 0.4196 | Yes | ||
| 8 | SOD1 | 1435304_at 1440222_at 1440896_at 1451124_at 1459976_s_at | 593 | 2.610 | 0.4637 | Yes | ||
| 9 | TMEM147 | 1428465_at | 657 | 2.466 | 0.5070 | Yes | ||
| 10 | GTF3A | 1427027_a_at | 933 | 2.048 | 0.5328 | Yes | ||
| 11 | YWHAQ | 1420828_s_at 1420829_a_at 1420830_x_at 1432842_s_at 1437608_x_at 1454378_at 1460590_s_at 1460621_x_at | 1495 | 1.636 | 0.5379 | Yes | ||
| 12 | MOAP1 | 1431823_at 1448787_at | 1626 | 1.544 | 0.5609 | Yes | ||
| 13 | CBX7 | 1420039_s_at 1433557_at 1449713_at | 2168 | 1.284 | 0.5602 | Yes | ||
| 14 | YY1 | 1422569_at 1422570_at 1435824_at 1457834_at | 2504 | 1.154 | 0.5665 | Yes | ||
| 15 | PSMD8 | 1423296_at | 2644 | 1.117 | 0.5811 | Yes | ||
| 16 | MRPL40 | 1448849_at | 4550 | 0.718 | 0.5076 | No | ||
| 17 | PLCL2 | 1426450_at | 4669 | 0.695 | 0.5152 | No | ||
| 18 | PFN2 | 1418209_a_at 1418210_at 1436993_x_at | 5132 | 0.618 | 0.5057 | No | ||
| 19 | PFKP | 1416069_at 1430634_a_at 1437759_at | 5588 | 0.554 | 0.4953 | No | ||
| 20 | DDX19A | 1417927_at | 5723 | 0.537 | 0.4992 | No | ||
| 21 | PFTK1 | 1419249_at 1419250_a_at 1441692_at 1445984_at 1453956_a_at | 7120 | 0.373 | 0.4425 | No | ||
| 22 | DCTN3 | 1416247_at | 7580 | 0.324 | 0.4276 | No | ||
| 23 | MYLK | 1425504_at 1425505_at 1425506_at | 9799 | 0.119 | 0.3285 | No | ||
| 24 | ZHX2 | 1455210_at | 9967 | 0.104 | 0.3228 | No | ||
| 25 | MAGED2 | 1426306_a_at 1459873_x_at | 10380 | 0.070 | 0.3053 | No | ||
| 26 | PPP1R2 | 1417341_a_at 1417342_at 1448684_at 1456830_at | 11027 | 0.014 | 0.2761 | No | ||
| 27 | SEP15 | 1448903_at 1454525_at | 12826 | -0.146 | 0.1967 | No | ||
| 28 | TSPYL4 | 1424029_at | 14104 | -0.267 | 0.1434 | No | ||
| 29 | HSPA4 | 1416146_at 1416147_at 1440575_at | 15600 | -0.430 | 0.0832 | No | ||
| 30 | PBX3 | 1421193_a_at 1440154_at 1442270_at 1447640_s_at | 15773 | -0.451 | 0.0838 | No | ||
| 31 | GLO1 | 1424108_at 1424109_a_at 1436070_at 1451240_a_at | 16158 | -0.500 | 0.0756 | No | ||
| 32 | FKBP1B | 1449429_at | 16365 | -0.526 | 0.0761 | No | ||
| 33 | LDOC1 | 1435815_at | 18053 | -0.809 | 0.0142 | No | ||
| 34 | UBE2D2 | 1416475_at 1416476_a_at 1416477_at 1439655_at 1445885_at 1448356_at | 19318 | -1.083 | -0.0232 | No | ||
| 35 | HNRPU | 1423050_s_at 1423051_at 1434390_at 1434391_at 1450849_at | 19626 | -1.172 | -0.0153 | No | ||
| 36 | NDUFC1 | 1416285_at 1448284_a_at | 19814 | -1.240 | -0.0006 | No | ||
| 37 | TMED10 | 1424707_at 1424708_at | 19984 | -1.303 | 0.0161 | No | ||
| 38 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 20737 | -1.715 | 0.0139 | No | ||
| 39 | PARP4 | 1441026_at | 21302 | -2.178 | 0.0290 | No |