Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
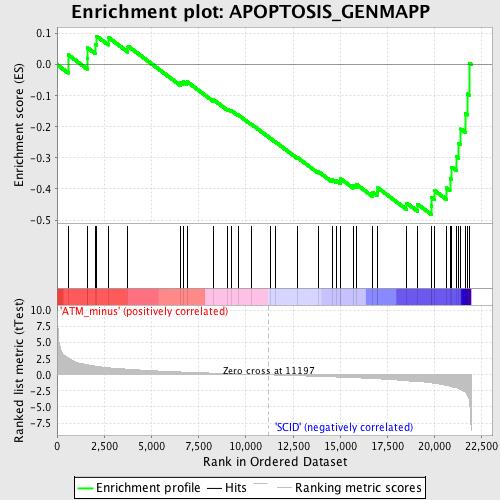
| GeneSet | APOPTOSIS_GENMAPP |
| Enrichment Score (ES) | -0.48210096 |
| Normalized Enrichment Score (NES) | -1.6319449 |
| Nominal p-value | 0.013071896 |
| FDR q-value | 0.36016718 |
| FWER p-Value | 1.0 |

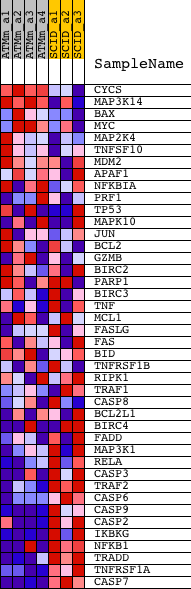
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CYCS | 1422484_at 1445484_at | 587 | 2.631 | 0.0319 | No | ||
| 2 | MAP3K14 | 1422999_at 1434364_at | 1607 | 1.556 | 0.0201 | No | ||
| 3 | BAX | 1416837_at | 1621 | 1.549 | 0.0541 | No | ||
| 4 | MYC | 1424942_a_at | 2017 | 1.346 | 0.0660 | No | ||
| 5 | MAP2K4 | 1426233_at 1442679_at 1451982_at | 2090 | 1.312 | 0.0920 | No | ||
| 6 | TNFSF10 | 1420412_at 1439680_at 1459913_at | 2718 | 1.094 | 0.0878 | No | ||
| 7 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 3752 | 0.867 | 0.0600 | No | ||
| 8 | APAF1 | 1450223_at | 6539 | 0.437 | -0.0575 | No | ||
| 9 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 6670 | 0.421 | -0.0541 | No | ||
| 10 | PRF1 | 1451862_a_at | 6879 | 0.400 | -0.0547 | No | ||
| 11 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 8278 | 0.258 | -0.1127 | No | ||
| 12 | MAPK10 | 1437195_x_at 1448342_at 1458002_at 1458062_at 1458513_at | 9031 | 0.187 | -0.1429 | No | ||
| 13 | JUN | 1417409_at 1448694_at | 9216 | 0.171 | -0.1475 | No | ||
| 14 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 9591 | 0.137 | -0.1615 | No | ||
| 15 | GZMB | 1419060_at | 10282 | 0.079 | -0.1913 | No | ||
| 16 | BIRC2 | 1418854_at | 11288 | -0.008 | -0.2370 | No | ||
| 17 | PARP1 | 1422502_at 1422503_s_at 1435368_a_at 1443573_at | 11562 | -0.034 | -0.2487 | No | ||
| 18 | BIRC3 | 1421392_a_at 1425223_at | 12726 | -0.137 | -0.2988 | No | ||
| 19 | TNF | 1419607_at | 13820 | -0.238 | -0.3434 | No | ||
| 20 | MCL1 | 1416880_at 1416881_at 1437527_x_at 1448503_at 1456243_x_at 1456381_x_at | 14568 | -0.313 | -0.3705 | No | ||
| 21 | FASLG | 1418803_a_at 1449235_at | 14775 | -0.338 | -0.3724 | No | ||
| 22 | FAS | 1460251_at | 15007 | -0.363 | -0.3748 | No | ||
| 23 | BID | 1417045_at 1447873_x_at 1448560_at | 15017 | -0.364 | -0.3671 | No | ||
| 24 | TNFRSF1B | 1418099_at 1448951_at | 15672 | -0.439 | -0.3872 | No | ||
| 25 | RIPK1 | 1419508_at 1439273_at 1449485_at | 15871 | -0.462 | -0.3859 | No | ||
| 26 | TRAF1 | 1423602_at 1445452_at | 16705 | -0.577 | -0.4111 | No | ||
| 27 | CASP8 | 1424552_at | 16947 | -0.612 | -0.4085 | No | ||
| 28 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 16991 | -0.619 | -0.3966 | No | ||
| 29 | BIRC4 | 1421394_a_at 1426636_a_at 1437533_at 1440351_at 1450231_a_at 1450232_at 1456088_at | 18513 | -0.924 | -0.4455 | No | ||
| 30 | FADD | 1416888_at 1446499_at | 19092 | -1.031 | -0.4489 | No | ||
| 31 | MAP3K1 | 1424850_at | 19820 | -1.242 | -0.4544 | Yes | ||
| 32 | RELA | 1419536_a_at | 19848 | -1.253 | -0.4277 | Yes | ||
| 33 | CASP3 | 1426165_a_at 1430192_at 1449839_at | 19993 | -1.310 | -0.4050 | Yes | ||
| 34 | TRAF2 | 1451233_at | 20609 | -1.634 | -0.3967 | Yes | ||
| 35 | CASP6 | 1415995_at 1458715_at | 20811 | -1.766 | -0.3665 | Yes | ||
| 36 | CASP9 | 1426125_a_at 1437537_at 1457816_at | 20906 | -1.849 | -0.3295 | Yes | ||
| 37 | CASP2 | 1448165_at | 21150 | -2.026 | -0.2954 | Yes | ||
| 38 | IKBKG | 1421208_at 1421209_s_at 1435646_at 1435647_at 1450161_at 1454690_at | 21274 | -2.150 | -0.2531 | Yes | ||
| 39 | NFKB1 | 1427705_a_at 1442949_at | 21385 | -2.296 | -0.2069 | Yes | ||
| 40 | TRADD | 1429117_at 1439910_a_at 1452622_a_at | 21619 | -2.690 | -0.1575 | Yes | ||
| 41 | TNFRSF1A | 1417291_at | 21718 | -3.039 | -0.0942 | Yes | ||
| 42 | CASP7 | 1426062_a_at 1448659_at | 21858 | -4.665 | 0.0036 | Yes |