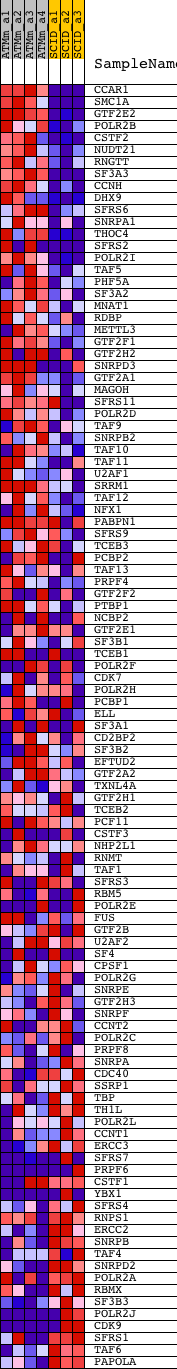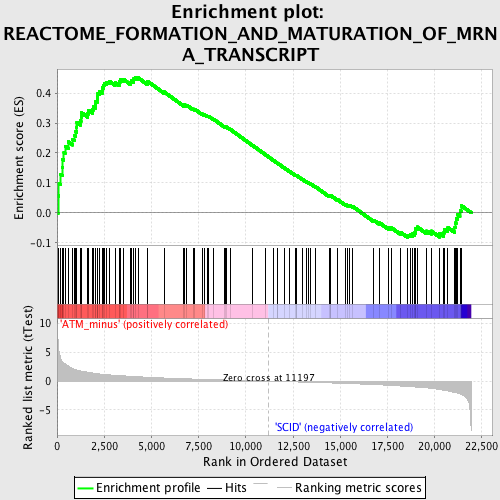
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_FORMATION_AND_MATURATION_OF_MRNA_TRANSCRIPT |
| Enrichment Score (ES) | 0.4517522 |
| Normalized Enrichment Score (NES) | 1.684835 |
| Nominal p-value | 0.0016891892 |
| FDR q-value | 0.43104932 |
| FWER p-Value | 0.97 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCAR1 | 1436156_at 1436157_at 1453319_at | 49 | 7.222 | 0.0581 | Yes | ||
| 2 | SMC1A | 1417830_at 1417831_at 1417832_at | 96 | 5.132 | 0.0988 | Yes | ||
| 3 | GTF2E2 | 1448884_at | 195 | 3.902 | 0.1269 | Yes | ||
| 4 | POLR2B | 1433552_a_at | 272 | 3.375 | 0.1517 | Yes | ||
| 5 | CSTF2 | 1419644_at 1419645_at 1455523_at | 289 | 3.323 | 0.1787 | Yes | ||
| 6 | NUDT21 | 1417681_at 1428339_at 1437213_at 1455966_s_at | 361 | 3.083 | 0.2012 | Yes | ||
| 7 | RNGTT | 1425844_a_at 1448714_at | 423 | 2.974 | 0.2232 | Yes | ||
| 8 | SF3A3 | 1423811_at | 594 | 2.610 | 0.2372 | Yes | ||
| 9 | CCNH | 1418585_at | 829 | 2.208 | 0.2450 | Yes | ||
| 10 | DHX9 | 1425617_at 1425618_at 1451770_s_at | 916 | 2.072 | 0.2583 | Yes | ||
| 11 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 990 | 1.984 | 0.2716 | Yes | ||
| 12 | SNRPA1 | 1417351_a_at 1417352_s_at 1417353_x_at | 1037 | 1.942 | 0.2857 | Yes | ||
| 13 | THOC4 | 1417724_at 1436211_at | 1047 | 1.933 | 0.3014 | Yes | ||
| 14 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 1230 | 1.777 | 0.3079 | Yes | ||
| 15 | POLR2I | 1428494_a_at | 1269 | 1.751 | 0.3208 | Yes | ||
| 16 | TAF5 | 1435474_at | 1291 | 1.741 | 0.3344 | Yes | ||
| 17 | PHF5A | 1424170_at | 1625 | 1.545 | 0.3320 | Yes | ||
| 18 | SF3A2 | 1450576_a_at 1455546_s_at | 1672 | 1.526 | 0.3427 | Yes | ||
| 19 | MNAT1 | 1432177_a_at | 1885 | 1.414 | 0.3448 | Yes | ||
| 20 | RDBP | 1450705_at | 1918 | 1.397 | 0.3550 | Yes | ||
| 21 | METTL3 | 1423099_a_at | 2032 | 1.341 | 0.3610 | Yes | ||
| 22 | GTF2F1 | 1417698_at 1417699_at | 2047 | 1.333 | 0.3715 | Yes | ||
| 23 | GTF2H2 | 1450701_a_at 1457818_at | 2119 | 1.302 | 0.3791 | Yes | ||
| 24 | SNRPD3 | 1422884_at 1422885_at 1449629_s_at | 2144 | 1.292 | 0.3888 | Yes | ||
| 25 | GTF2A1 | 1421357_at | 2158 | 1.287 | 0.3990 | Yes | ||
| 26 | MAGOH | 1416212_at | 2254 | 1.242 | 0.4050 | Yes | ||
| 27 | SFRS11 | 1427269_at 1430077_at 1439095_at 1452371_at | 2384 | 1.197 | 0.4091 | Yes | ||
| 28 | POLR2D | 1424258_at | 2408 | 1.187 | 0.4179 | Yes | ||
| 29 | TAF9 | 1422778_at 1451509_at | 2462 | 1.167 | 0.4253 | Yes | ||
| 30 | SNRPB2 | 1426613_a_at 1452422_a_at | 2492 | 1.158 | 0.4336 | Yes | ||
| 31 | TAF10 | 1448784_at | 2626 | 1.121 | 0.4369 | Yes | ||
| 32 | TAF11 | 1446161_at 1451995_at | 2765 | 1.083 | 0.4396 | Yes | ||
| 33 | U2AF1 | 1422509_at | 3069 | 1.010 | 0.4342 | Yes | ||
| 34 | SRRM1 | 1420934_a_at 1420935_a_at 1442330_at 1447447_s_at 1450045_at 1454689_at 1459828_at | 3295 | 0.966 | 0.4319 | Yes | ||
| 35 | TAF12 | 1423398_at | 3310 | 0.963 | 0.4393 | Yes | ||
| 36 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 3364 | 0.951 | 0.4448 | Yes | ||
| 37 | PABPN1 | 1415728_at 1422848_a_at 1422849_a_at 1422850_at | 3492 | 0.924 | 0.4467 | Yes | ||
| 38 | SFRS9 | 1417727_at | 3882 | 0.842 | 0.4359 | Yes | ||
| 39 | TCEB3 | 1422591_at 1434117_at 1450676_at | 3922 | 0.834 | 0.4411 | Yes | ||
| 40 | PCBP2 | 1421743_a_at 1435881_at | 4061 | 0.808 | 0.4415 | Yes | ||
| 41 | TAF13 | 1448707_at | 4066 | 0.808 | 0.4481 | Yes | ||
| 42 | PRPF4 | 1429724_at 1438481_at | 4132 | 0.794 | 0.4518 | Yes | ||
| 43 | GTF2F2 | 1426626_at 1443233_at | 4284 | 0.767 | 0.4512 | No | ||
| 44 | PTBP1 | 1424874_a_at 1450443_at 1458284_at | 4773 | 0.679 | 0.4346 | No | ||
| 45 | NCBP2 | 1423045_at 1423046_s_at 1450847_at | 4790 | 0.676 | 0.4395 | No | ||
| 46 | GTF2E1 | 1428124_at 1443110_at | 5666 | 0.544 | 0.4039 | No | ||
| 47 | SF3B1 | 1418561_at 1418562_at 1440818_s_at 1449138_at | 6715 | 0.418 | 0.3594 | No | ||
| 48 | TCEB1 | 1427914_a_at 1427915_s_at 1436226_at 1452593_a_at | 6752 | 0.414 | 0.3612 | No | ||
| 49 | POLR2F | 1415754_at 1427402_at | 6862 | 0.402 | 0.3596 | No | ||
| 50 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 7205 | 0.362 | 0.3469 | No | ||
| 51 | POLR2H | 1424473_at | 7291 | 0.354 | 0.3460 | No | ||
| 52 | PCBP1 | 1448642_at | 7692 | 0.315 | 0.3303 | No | ||
| 53 | ELL | 1441928_x_at 1460643_at | 7782 | 0.306 | 0.3288 | No | ||
| 54 | SF3A1 | 1419087_s_at 1445719_at 1446687_at 1449333_at | 7965 | 0.290 | 0.3229 | No | ||
| 55 | CD2BP2 | 1417223_at 1417224_a_at 1448624_at | 8011 | 0.286 | 0.3232 | No | ||
| 56 | SF3B2 | 1429362_a_at 1433461_at 1438637_x_at 1453123_at 1456040_at 1456352_a_at 1459836_x_at | 8287 | 0.257 | 0.3128 | No | ||
| 57 | EFTUD2 | 1416557_a_at 1438835_a_at 1438836_at 1456107_x_at | 8890 | 0.200 | 0.2869 | No | ||
| 58 | GTF2A2 | 1433528_at | 8934 | 0.195 | 0.2865 | No | ||
| 59 | TXNL4A | 1419179_at | 8958 | 0.193 | 0.2871 | No | ||
| 60 | GTF2H1 | 1453169_a_at | 9163 | 0.176 | 0.2792 | No | ||
| 61 | TCEB2 | 1428263_a_at | 10344 | 0.073 | 0.2258 | No | ||
| 62 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 11052 | 0.012 | 0.1935 | No | ||
| 63 | CSTF3 | 1424723_s_at 1443909_at | 11465 | -0.025 | 0.1748 | No | ||
| 64 | NHP2L1 | 1416973_at | 11664 | -0.044 | 0.1661 | No | ||
| 65 | RNMT | 1436328_at 1440002_at 1446442_at 1453106_a_at | 11665 | -0.044 | 0.1665 | No | ||
| 66 | TAF1 | 1447417_at 1455513_at 1458744_at | 12022 | -0.074 | 0.1508 | No | ||
| 67 | SFRS3 | 1416150_a_at 1416151_at 1416152_a_at 1454993_a_at | 12329 | -0.101 | 0.1376 | No | ||
| 68 | RBM5 | 1438069_a_at 1441319_at 1444618_at 1444971_at 1452186_at 1452187_at 1456262_at | 12628 | -0.128 | 0.1250 | No | ||
| 69 | POLR2E | 1417138_s_at 1451093_at 1458326_at | 12659 | -0.131 | 0.1247 | No | ||
| 70 | FUS | 1451285_at 1451286_s_at 1455831_at 1458801_at | 12690 | -0.134 | 0.1245 | No | ||
| 71 | GTF2B | 1438066_at 1451135_at | 12992 | -0.163 | 0.1121 | No | ||
| 72 | U2AF2 | 1417260_at | 13184 | -0.180 | 0.1048 | No | ||
| 73 | SF4 | 1424381_at 1444699_at | 13312 | -0.191 | 0.1006 | No | ||
| 74 | CPSF1 | 1417665_a_at | 13443 | -0.203 | 0.0963 | No | ||
| 75 | POLR2G | 1416270_at | 13673 | -0.223 | 0.0877 | No | ||
| 76 | SNRPE | 1451294_s_at | 14431 | -0.299 | 0.0555 | No | ||
| 77 | GTF2H3 | 1454092_a_at | 14466 | -0.303 | 0.0565 | No | ||
| 78 | SNRPF | 1428672_at | 14478 | -0.305 | 0.0585 | No | ||
| 79 | CCNT2 | 1427088_at 1427089_at 1435445_at 1452297_at 1453399_at 1457439_at | 14874 | -0.348 | 0.0433 | No | ||
| 80 | POLR2C | 1416341_at | 15287 | -0.393 | 0.0278 | No | ||
| 81 | PRPF8 | 1422453_at | 15399 | -0.406 | 0.0261 | No | ||
| 82 | SNRPA | 1417274_at | 15500 | -0.417 | 0.0250 | No | ||
| 83 | CDC40 | 1442667_at 1445348_at 1453342_at 1457366_at | 15645 | -0.436 | 0.0220 | No | ||
| 84 | SSRP1 | 1426788_a_at 1426789_s_at 1426790_at | 16776 | -0.586 | -0.0249 | No | ||
| 85 | TBP | 1426469_a_at 1426470_at | 17082 | -0.633 | -0.0335 | No | ||
| 86 | TH1L | 1416198_at | 17571 | -0.716 | -0.0499 | No | ||
| 87 | POLR2L | 1428296_at | 17711 | -0.740 | -0.0501 | No | ||
| 88 | CCNT1 | 1419313_at 1456477_at | 18208 | -0.852 | -0.0657 | No | ||
| 89 | ERCC3 | 1448497_at | 18568 | -0.937 | -0.0743 | No | ||
| 90 | SFRS7 | 1424033_at 1424883_s_at 1436871_at | 18695 | -0.964 | -0.0721 | No | ||
| 91 | PRPF6 | 1424036_at 1426026_at 1454789_x_at | 18836 | -0.989 | -0.0702 | No | ||
| 92 | CSTF1 | 1417117_at 1448597_at | 18947 | -1.008 | -0.0668 | No | ||
| 93 | YBX1 | 1418624_at 1430720_at 1438064_at | 18974 | -1.012 | -0.0596 | No | ||
| 94 | SFRS4 | 1448778_at | 18986 | -1.014 | -0.0516 | No | ||
| 95 | RNPS1 | 1437027_x_at 1437359_at 1437851_x_at 1438267_x_at 1448324_at | 19075 | -1.028 | -0.0470 | No | ||
| 96 | ERCC2 | 1460727_at | 19582 | -1.158 | -0.0605 | No | ||
| 97 | SNRPB | 1419260_a_at 1437193_s_at | 19808 | -1.239 | -0.0605 | No | ||
| 98 | TAF4 | 1427460_at 1452438_s_at | 20254 | -1.435 | -0.0689 | No | ||
| 99 | SNRPD2 | 1452680_at | 20486 | -1.550 | -0.0665 | No | ||
| 100 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 20505 | -1.561 | -0.0543 | No | ||
| 101 | RBMX | 1416177_at 1416354_at 1416355_at 1426863_at 1437847_x_at | 20699 | -1.696 | -0.0490 | No | ||
| 102 | SF3B3 | 1415985_at 1430075_at | 21067 | -1.964 | -0.0494 | No | ||
| 103 | POLR2J | 1417720_at 1440065_at | 21085 | -1.978 | -0.0337 | No | ||
| 104 | CDK9 | 1417269_at 1457240_at | 21134 | -2.017 | -0.0190 | No | ||
| 105 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 21222 | -2.084 | -0.0056 | No | ||
| 106 | TAF6 | 1418593_at | 21339 | -2.227 | 0.0077 | No | ||
| 107 | PAPOLA | 1424216_a_at 1424217_at 1427544_a_at 1444015_at 1455836_at | 21416 | -2.348 | 0.0238 | No |