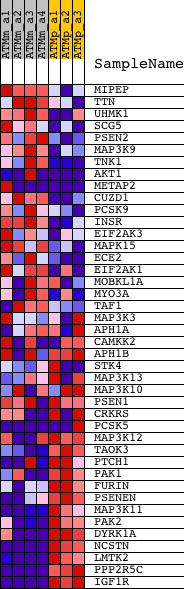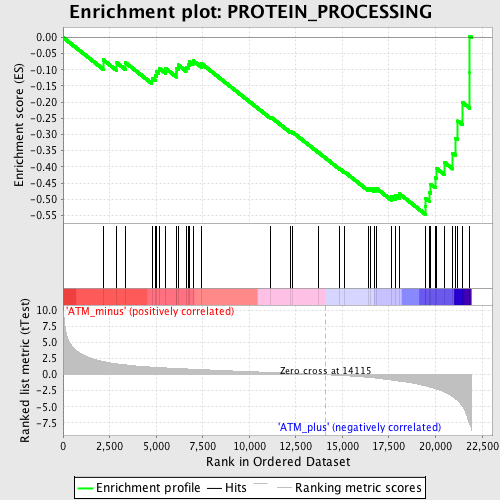
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | PROTEIN_PROCESSING |
| Enrichment Score (ES) | -0.54711956 |
| Normalized Enrichment Score (NES) | -1.75521 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.24928151 |
| FWER p-Value | 0.891 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MIPEP | 1428290_at 1459294_at | 2153 | 2.037 | -0.0692 | No | ||
| 2 | TTN | 1427445_a_at 1427446_s_at 1431928_at 1444083_at 1444638_at 1446450_at | 2890 | 1.686 | -0.0787 | No | ||
| 3 | UHMK1 | 1446120_at 1454260_at | 3352 | 1.526 | -0.0779 | No | ||
| 4 | SCG5 | 1423150_at 1446755_at | 4773 | 1.177 | -0.1260 | No | ||
| 5 | PSEN2 | 1425869_a_at 1431542_at | 4952 | 1.143 | -0.1177 | No | ||
| 6 | MAP3K9 | 1432981_at 1442218_at 1455684_at 1458922_at | 5022 | 1.128 | -0.1048 | No | ||
| 7 | TNK1 | 1422721_at 1425634_a_at 1425635_at | 5149 | 1.108 | -0.0947 | No | ||
| 8 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 5518 | 1.047 | -0.0965 | No | ||
| 9 | METAP2 | 1423610_at 1434120_a_at 1436531_at 1447263_at 1451048_at 1457416_at | 6085 | 0.974 | -0.1084 | No | ||
| 10 | CUZD1 | 1417413_at | 6097 | 0.973 | -0.0950 | No | ||
| 11 | PCSK9 | 1437453_s_at | 6179 | 0.960 | -0.0850 | No | ||
| 12 | INSR | 1421380_at 1450225_at | 6601 | 0.897 | -0.0914 | No | ||
| 13 | EIF2AK3 | 1430371_x_at 1449278_at 1453505_a_at | 6713 | 0.883 | -0.0838 | No | ||
| 14 | MAPK15 | 1441755_at | 6803 | 0.870 | -0.0755 | No | ||
| 15 | ECE2 | 1424561_at | 6978 | 0.843 | -0.0714 | No | ||
| 16 | EIF2AK1 | 1421900_at 1421901_at 1430696_at 1436006_at 1453902_at | 7432 | 0.783 | -0.0808 | No | ||
| 17 | MOBKL1A | 1430564_at 1455184_at | 11163 | 0.345 | -0.2463 | No | ||
| 18 | MYO3A | 1431983_at | 12205 | 0.231 | -0.2905 | No | ||
| 19 | TAF1 | 1447417_at 1455513_at 1458744_at | 12317 | 0.219 | -0.2925 | No | ||
| 20 | MAP3K3 | 1426686_s_at 1426687_at 1436522_at | 13720 | 0.051 | -0.3558 | No | ||
| 21 | APH1A | 1424979_at 1424980_s_at 1451554_a_at | 14845 | -0.113 | -0.4055 | No | ||
| 22 | CAMKK2 | 1424474_a_at 1424475_at 1424476_at 1455401_at | 15135 | -0.159 | -0.4164 | No | ||
| 23 | APH1B | 1435793_at 1456500_at | 16384 | -0.416 | -0.4675 | No | ||
| 24 | STK4 | 1421107_at 1434159_at 1436015_s_at | 16530 | -0.452 | -0.4676 | No | ||
| 25 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 16696 | -0.498 | -0.4681 | No | ||
| 26 | MAP3K10 | 1436373_at 1440500_at 1457456_at 1458651_at | 16842 | -0.543 | -0.4669 | No | ||
| 27 | PSEN1 | 1420261_at 1421853_at 1425549_at 1450399_at | 17630 | -0.831 | -0.4910 | No | ||
| 28 | CRKRS | 1425555_at 1425556_at 1438831_at 1440858_at 1443294_at 1455567_at 1455964_at | 17841 | -0.926 | -0.4873 | No | ||
| 29 | PCSK5 | 1424605_at 1437339_s_at 1438248_at 1451406_a_at 1452475_at 1459124_at | 18053 | -0.999 | -0.4827 | No | ||
| 30 | MAP3K12 | 1417115_at 1438908_at 1456565_s_at | 19465 | -1.743 | -0.5222 | Yes | ||
| 31 | TAOK3 | 1435964_a_at 1444366_at 1444881_at 1447257_at 1455733_at | 19478 | -1.757 | -0.4976 | Yes | ||
| 32 | PTCH1 | 1428853_at 1439663_at 1447039_at 1450824_at | 19675 | -1.934 | -0.4789 | Yes | ||
| 33 | PAK1 | 1420979_at 1420980_at 1450070_s_at 1459029_at | 19738 | -1.986 | -0.4533 | Yes | ||
| 34 | FURIN | 1418518_at | 19994 | -2.174 | -0.4339 | Yes | ||
| 35 | PSENEN | 1415679_at | 20049 | -2.219 | -0.4046 | Yes | ||
| 36 | MAP3K11 | 1450669_at | 20460 | -2.667 | -0.3852 | Yes | ||
| 37 | PAK2 | 1434250_at 1454887_at | 20929 | -3.442 | -0.3573 | Yes | ||
| 38 | DYRK1A | 1426226_at 1428299_at 1451977_at | 21090 | -3.760 | -0.3109 | Yes | ||
| 39 | NCSTN | 1418570_at 1444532_at | 21196 | -4.020 | -0.2582 | Yes | ||
| 40 | LMTK2 | 1439871_at 1455018_at 1459531_at | 21433 | -4.820 | -0.2000 | Yes | ||
| 41 | PPP2R5C | 1425542_a_at 1425725_s_at 1425726_x_at 1427003_at 1434205_at 1434206_s_at 1444828_at | 21843 | -7.790 | -0.1073 | Yes | ||
| 42 | IGF1R | 1426565_at 1437219_at 1443566_at 1445600_at 1445664_at 1446303_at | 21847 | -7.797 | 0.0041 | Yes |