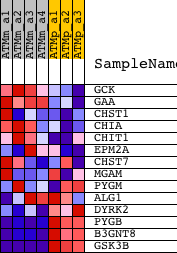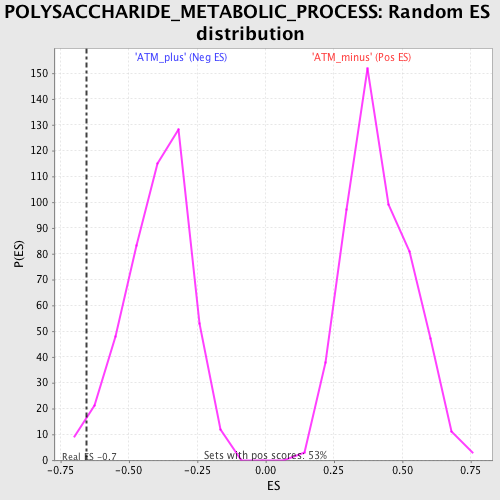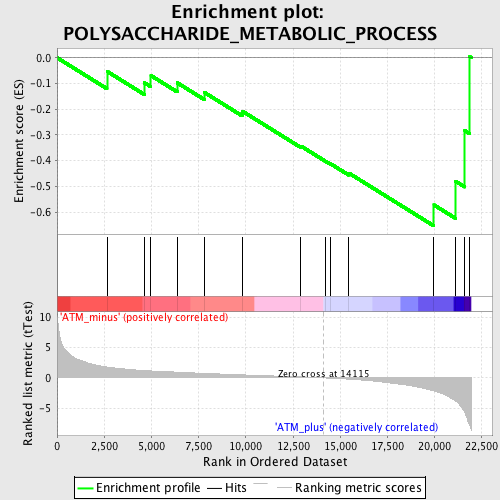
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | POLYSACCHARIDE_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.65396583 |
| Normalized Enrichment Score (NES) | -1.6496524 |
| Nominal p-value | 0.019189766 |
| FDR q-value | 0.32577735 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GCK | 1419146_a_at 1425303_at | 2643 | 1.772 | -0.0529 | No | ||
| 2 | GAA | 1419428_a_at 1436849_x_at | 4635 | 1.205 | -0.0977 | No | ||
| 3 | CHST1 | 1449147_at | 4971 | 1.138 | -0.0696 | No | ||
| 4 | CHIA | 1416456_a_at | 6362 | 0.932 | -0.0974 | No | ||
| 5 | CHIT1 | 1453844_at | 7817 | 0.735 | -0.1356 | No | ||
| 6 | EPM2A | 1421553_at 1439677_at | 9801 | 0.493 | -0.2073 | No | ||
| 7 | CHST7 | 1449402_at | 12907 | 0.151 | -0.3431 | No | ||
| 8 | MGAM | 1456777_at | 14219 | -0.017 | -0.4023 | No | ||
| 9 | PYGM | 1448602_at | 14465 | -0.052 | -0.4114 | No | ||
| 10 | ALG1 | 1451368_at | 15445 | -0.210 | -0.4481 | No | ||
| 11 | DYRK2 | 1428637_at 1436918_at 1456502_at 1457667_x_at | 19960 | -2.139 | -0.5723 | Yes | ||
| 12 | PYGB | 1433504_at | 21094 | -3.780 | -0.4796 | Yes | ||
| 13 | B3GNT8 | 1425128_at | 21596 | -5.732 | -0.2836 | Yes | ||
| 14 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 21861 | -7.833 | 0.0034 | Yes |