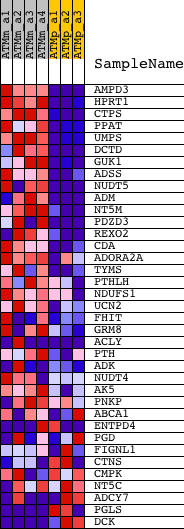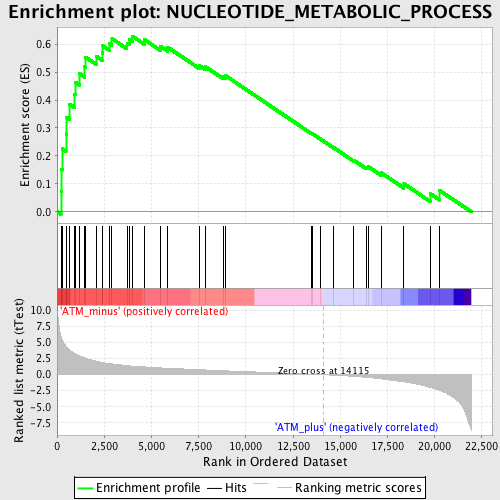
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | NUCLEOTIDE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.62860113 |
| Normalized Enrichment Score (NES) | 1.9456276 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.018175831 |
| FWER p-Value | 0.078 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AMPD3 | 1422573_at | 207 | 5.889 | 0.0733 | Yes | ||
| 2 | HPRT1 | 1448736_a_at | 235 | 5.625 | 0.1511 | Yes | ||
| 3 | CTPS | 1416563_at | 268 | 5.392 | 0.2254 | Yes | ||
| 4 | PPAT | 1428543_at 1441770_at 1452831_s_at | 473 | 4.443 | 0.2785 | Yes | ||
| 5 | UMPS | 1431652_at 1434859_at 1451445_at 1455832_a_at | 517 | 4.296 | 0.3369 | Yes | ||
| 6 | DCTD | 1454659_at | 659 | 3.888 | 0.3851 | Yes | ||
| 7 | GUK1 | 1416395_at | 915 | 3.314 | 0.4200 | Yes | ||
| 8 | ADSS | 1460726_at | 954 | 3.247 | 0.4639 | Yes | ||
| 9 | NUDT5 | 1430341_at 1448651_at | 1187 | 2.916 | 0.4943 | Yes | ||
| 10 | ADM | 1416077_at 1447839_x_at | 1462 | 2.631 | 0.5188 | Yes | ||
| 11 | NT5M | 1453767_a_at | 1506 | 2.588 | 0.5532 | Yes | ||
| 12 | PDZD3 | 1449330_at | 2093 | 2.082 | 0.5557 | Yes | ||
| 13 | REXO2 | 1451259_at | 2395 | 1.895 | 0.5685 | Yes | ||
| 14 | CDA | 1427357_at | 2417 | 1.876 | 0.5939 | Yes | ||
| 15 | ADORA2A | 1427519_at 1460710_at | 2763 | 1.730 | 0.6025 | Yes | ||
| 16 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 2905 | 1.680 | 0.6197 | Yes | ||
| 17 | PTHLH | 1422324_a_at 1427527_a_at | 3701 | 1.409 | 0.6032 | Yes | ||
| 18 | NDUFS1 | 1425143_a_at 1453354_at | 3836 | 1.373 | 0.6163 | Yes | ||
| 19 | UCN2 | 1450607_s_at | 3980 | 1.338 | 0.6286 | Yes | ||
| 20 | FHIT | 1425893_a_at | 4609 | 1.211 | 0.6169 | No | ||
| 21 | GRM8 | 1421530_a_at | 5490 | 1.050 | 0.5915 | No | ||
| 22 | ACLY | 1425326_at 1438389_x_at 1439445_x_at 1439459_x_at 1446315_at 1451666_at | 5861 | 1.002 | 0.5887 | No | ||
| 23 | PTH | 1419345_at | 7535 | 0.770 | 0.5231 | No | ||
| 24 | ADK | 1416319_at 1421767_at 1438292_x_at 1442615_at 1445402_at 1446068_at 1446675_at 1456960_at 1459645_at | 7837 | 0.731 | 0.5197 | No | ||
| 25 | NUDT4 | 1418505_at 1449107_at | 8802 | 0.610 | 0.4842 | No | ||
| 26 | AK5 | 1426339_at 1457032_at | 8936 | 0.594 | 0.4865 | No | ||
| 27 | PNKP | 1416378_at | 13479 | 0.084 | 0.2803 | No | ||
| 28 | ABCA1 | 1421839_at 1421840_at 1450392_at | 13514 | 0.080 | 0.2798 | No | ||
| 29 | ENTPD4 | 1431761_at 1438177_x_at 1447900_x_at 1449190_a_at | 13975 | 0.018 | 0.2591 | No | ||
| 30 | PGD | 1423706_a_at 1436771_x_at 1437380_x_at 1438627_x_at | 14658 | -0.084 | 0.2291 | No | ||
| 31 | FIGNL1 | 1422430_at 1460746_at | 15716 | -0.264 | 0.1846 | No | ||
| 32 | CTNS | 1416274_at 1456450_at | 16380 | -0.415 | 0.1601 | No | ||
| 33 | CMPK | 1423073_at 1440308_at | 16500 | -0.445 | 0.1609 | No | ||
| 34 | NT5C | 1417252_at | 17160 | -0.649 | 0.1400 | No | ||
| 35 | ADCY7 | 1420970_at 1450065_at 1456307_s_at | 18370 | -1.106 | 0.1003 | No | ||
| 36 | PGLS | 1417722_at 1457728_at | 19784 | -2.010 | 0.0640 | No | ||
| 37 | DCK | 1428838_a_at 1439012_a_at 1449176_a_at | 20266 | -2.433 | 0.0763 | No |