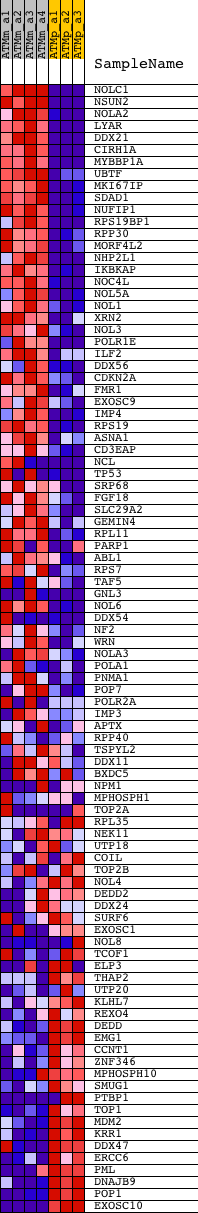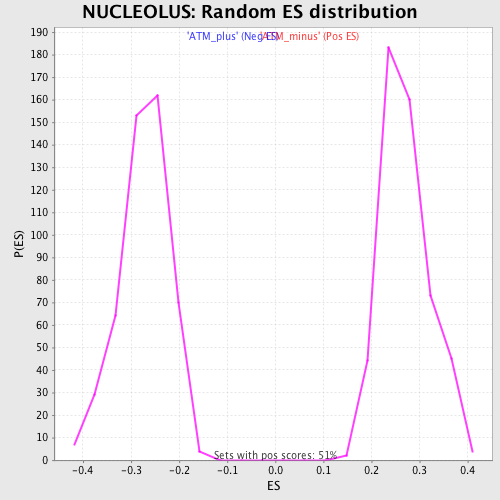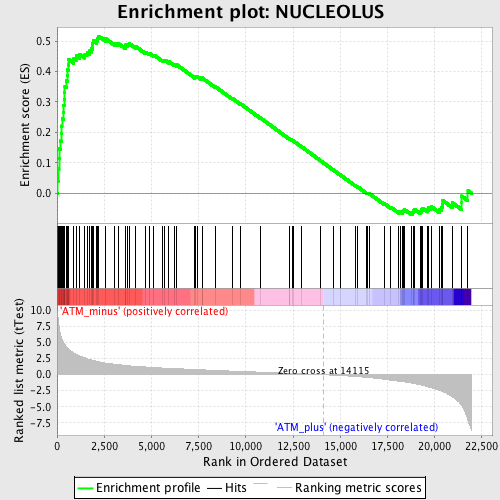
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | NUCLEOLUS |
| Enrichment Score (ES) | 0.5164936 |
| Normalized Enrichment Score (NES) | 1.9066253 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.01657746 |
| FWER p-Value | 0.153 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NOLC1 | 1428869_at 1428870_at 1450087_a_at | 33 | 9.043 | 0.0405 | Yes | ||
| 2 | NSUN2 | 1423850_at 1453759_at | 59 | 8.723 | 0.0799 | Yes | ||
| 3 | NOLA2 | 1416605_at 1416606_s_at | 100 | 7.580 | 0.1132 | Yes | ||
| 4 | LYAR | 1417511_at | 114 | 7.118 | 0.1457 | Yes | ||
| 5 | DDX21 | 1448270_at 1448271_a_at | 178 | 6.224 | 0.1717 | Yes | ||
| 6 | CIRH1A | 1423884_at | 234 | 5.643 | 0.1954 | Yes | ||
| 7 | MYBBP1A | 1423430_at 1423431_a_at | 240 | 5.541 | 0.2209 | Yes | ||
| 8 | UBTF | 1453097_a_at 1455490_at 1460304_a_at | 259 | 5.442 | 0.2454 | Yes | ||
| 9 | MKI67IP | 1424001_at | 334 | 5.045 | 0.2654 | Yes | ||
| 10 | SDAD1 | 1437088_at 1452454_at | 345 | 4.998 | 0.2882 | Yes | ||
| 11 | NUFIP1 | 1448802_at | 375 | 4.848 | 0.3094 | Yes | ||
| 12 | RPS19BP1 | 1455174_at | 389 | 4.788 | 0.3310 | Yes | ||
| 13 | RPP30 | 1423373_at | 414 | 4.705 | 0.3518 | Yes | ||
| 14 | MORF4L2 | 1415778_at 1439413_x_at | 480 | 4.420 | 0.3693 | Yes | ||
| 15 | NHP2L1 | 1416973_at | 534 | 4.229 | 0.3865 | Yes | ||
| 16 | IKBKAP | 1424142_at 1451254_at | 548 | 4.189 | 0.4054 | Yes | ||
| 17 | NOC4L | 1423826_at 1423827_s_at 1438095_x_at 1443794_x_at | 602 | 4.037 | 0.4217 | Yes | ||
| 18 | NOL5A | 1426533_at 1455035_s_at | 614 | 3.987 | 0.4397 | Yes | ||
| 19 | NOL1 | 1424019_at | 889 | 3.371 | 0.4428 | Yes | ||
| 20 | XRN2 | 1422842_at 1450777_at | 1006 | 3.163 | 0.4522 | Yes | ||
| 21 | NOL3 | 1444786_at 1451503_at | 1188 | 2.914 | 0.4575 | Yes | ||
| 22 | POLR1E | 1419058_at 1435057_x_at | 1474 | 2.617 | 0.4566 | Yes | ||
| 23 | ILF2 | 1417948_s_at 1417949_at | 1585 | 2.495 | 0.4631 | Yes | ||
| 24 | DDX56 | 1423815_at | 1725 | 2.364 | 0.4677 | Yes | ||
| 25 | CDKN2A | 1450140_a_at | 1811 | 2.297 | 0.4745 | Yes | ||
| 26 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 1865 | 2.245 | 0.4825 | Yes | ||
| 27 | EXOSC9 | 1418462_at | 1873 | 2.238 | 0.4926 | Yes | ||
| 28 | IMP4 | 1426213_at 1426214_at | 1900 | 2.224 | 0.5017 | Yes | ||
| 29 | RPS19 | 1449243_a_at 1460442_at | 2085 | 2.085 | 0.5030 | Yes | ||
| 30 | ASNA1 | 1418292_at | 2146 | 2.044 | 0.5097 | Yes | ||
| 31 | CD3EAP | 1430138_at | 2203 | 2.006 | 0.5165 | Yes | ||
| 32 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 2565 | 1.803 | 0.5083 | No | ||
| 33 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 3061 | 1.619 | 0.4932 | No | ||
| 34 | SRP68 | 1433708_at | 3240 | 1.556 | 0.4923 | No | ||
| 35 | FGF18 | 1449545_at | 3630 | 1.428 | 0.4811 | No | ||
| 36 | SLC29A2 | 1447748_x_at 1448257_at | 3637 | 1.427 | 0.4875 | No | ||
| 37 | GEMIN4 | 1433622_at | 3722 | 1.402 | 0.4901 | No | ||
| 38 | RPL11 | 1417615_a_at | 3812 | 1.378 | 0.4924 | No | ||
| 39 | PARP1 | 1422502_at 1422503_s_at 1435368_a_at 1443573_at | 4170 | 1.293 | 0.4821 | No | ||
| 40 | ABL1 | 1423999_at 1441291_at 1444134_at 1445153_at | 4702 | 1.192 | 0.4633 | No | ||
| 41 | RPS7 | 1435593_x_at | 4893 | 1.154 | 0.4600 | No | ||
| 42 | TAF5 | 1435474_at | 5130 | 1.111 | 0.4544 | No | ||
| 43 | GNL3 | 1433656_a_at 1440766_at | 5601 | 1.037 | 0.4377 | No | ||
| 44 | NOL6 | 1424472_at 1425629_a_at | 5705 | 1.023 | 0.4377 | No | ||
| 45 | DDX54 | 1438853_x_at 1447882_x_at 1460396_at | 5920 | 0.997 | 0.4325 | No | ||
| 46 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 6232 | 0.952 | 0.4227 | No | ||
| 47 | WRN | 1425074_at 1425982_a_at 1450163_a_at | 6346 | 0.935 | 0.4219 | No | ||
| 48 | NOLA3 | 1423210_a_at 1423211_at | 7298 | 0.800 | 0.3820 | No | ||
| 49 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 7331 | 0.795 | 0.3843 | No | ||
| 50 | PNMA1 | 1429224_at | 7443 | 0.782 | 0.3828 | No | ||
| 51 | POP7 | 1449162_at | 7675 | 0.752 | 0.3757 | No | ||
| 52 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 7677 | 0.752 | 0.3792 | No | ||
| 53 | IMP3 | 1416210_at | 8387 | 0.658 | 0.3498 | No | ||
| 54 | APTX | 1450921_at | 9289 | 0.553 | 0.3111 | No | ||
| 55 | RPP40 | 1424792_at 1443750_s_at | 9713 | 0.503 | 0.2941 | No | ||
| 56 | TSPYL2 | 1434849_at | 10757 | 0.389 | 0.2481 | No | ||
| 57 | DDX11 | 1438447_at 1441062_at | 12298 | 0.222 | 0.1787 | No | ||
| 58 | BXDC5 | 1428020_at 1431784_a_at 1459573_at | 12322 | 0.218 | 0.1786 | No | ||
| 59 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 12454 | 0.203 | 0.1736 | No | ||
| 60 | MPHOSPH1 | 1419792_at 1439695_a_at 1440924_at 1449612_x_at | 12510 | 0.198 | 0.1720 | No | ||
| 61 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 12942 | 0.148 | 0.1529 | No | ||
| 62 | RPL35 | 1416243_a_at 1434231_x_at 1436840_x_at 1454833_at 1454856_x_at 1455950_x_at | 13959 | 0.020 | 0.1065 | No | ||
| 63 | NEK11 | 1441525_at 1445012_at | 14662 | -0.084 | 0.0748 | No | ||
| 64 | UTP18 | 1447266_at 1454817_at | 14997 | -0.136 | 0.0601 | No | ||
| 65 | COIL | 1416692_at | 15796 | -0.280 | 0.0249 | No | ||
| 66 | TOP2B | 1416731_at 1416732_at 1448458_at 1456688_at | 15901 | -0.301 | 0.0215 | No | ||
| 67 | NOL4 | 1430189_at 1441447_at 1456387_at | 16406 | -0.421 | 0.0004 | No | ||
| 68 | DEDD2 | 1452070_at | 16449 | -0.430 | 0.0005 | No | ||
| 69 | DDX24 | 1415713_a_at 1441631_at | 16536 | -0.453 | -0.0014 | No | ||
| 70 | SURF6 | 1416864_at 1459331_at | 17329 | -0.703 | -0.0344 | No | ||
| 71 | EXOSC1 | 1432052_at 1452012_a_at 1453359_at | 17672 | -0.846 | -0.0461 | No | ||
| 72 | NOL8 | 1428950_s_at 1428951_at 1452974_at | 18097 | -1.010 | -0.0608 | No | ||
| 73 | TCOF1 | 1423600_a_at 1423601_s_at 1424643_at | 18195 | -1.037 | -0.0604 | No | ||
| 74 | ELP3 | 1426643_at 1436762_x_at 1437338_x_at 1443452_at | 18309 | -1.083 | -0.0606 | No | ||
| 75 | THAP2 | 1421177_at 1455336_at | 18334 | -1.095 | -0.0566 | No | ||
| 76 | UTP20 | 1452252_at 1459028_at | 18394 | -1.118 | -0.0541 | No | ||
| 77 | KLHL7 | 1428091_at 1452660_s_at | 18745 | -1.286 | -0.0642 | No | ||
| 78 | REXO4 | 1434113_a_at 1434114_at | 18849 | -1.346 | -0.0626 | No | ||
| 79 | DEDD | 1432000_a_at 1434994_at 1434995_s_at | 18890 | -1.369 | -0.0581 | No | ||
| 80 | EMG1 | 1416393_at | 18925 | -1.387 | -0.0532 | No | ||
| 81 | CCNT1 | 1419313_at 1456477_at | 19227 | -1.582 | -0.0596 | No | ||
| 82 | ZNF346 | 1417088_at 1448581_at | 19286 | -1.618 | -0.0548 | No | ||
| 83 | MPHOSPH10 | 1429080_at 1431388_at | 19342 | -1.657 | -0.0496 | No | ||
| 84 | SMUG1 | 1428763_at | 19613 | -1.885 | -0.0532 | No | ||
| 85 | PTBP1 | 1424874_a_at 1450443_at 1458284_at | 19680 | -1.938 | -0.0472 | No | ||
| 86 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 19804 | -2.024 | -0.0434 | No | ||
| 87 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 20268 | -2.435 | -0.0533 | No | ||
| 88 | KRR1 | 1420139_s_at 1428084_at 1444726_at 1452658_at | 20361 | -2.541 | -0.0457 | No | ||
| 89 | DDX47 | 1423752_at 1438380_at 1438381_x_at | 20407 | -2.611 | -0.0357 | No | ||
| 90 | ERCC6 | 1441595_at 1442604_at | 20428 | -2.634 | -0.0244 | No | ||
| 91 | PML | 1448757_at 1456103_at 1459137_at | 20925 | -3.435 | -0.0311 | No | ||
| 92 | DNAJB9 | 1417191_at | 21412 | -4.757 | -0.0313 | No | ||
| 93 | POP1 | 1428458_at 1441002_at | 21416 | -4.765 | -0.0093 | No | ||
| 94 | EXOSC10 | 1423587_a_at 1431832_x_at 1437294_at 1454021_a_at | 21761 | -7.113 | 0.0080 | No |