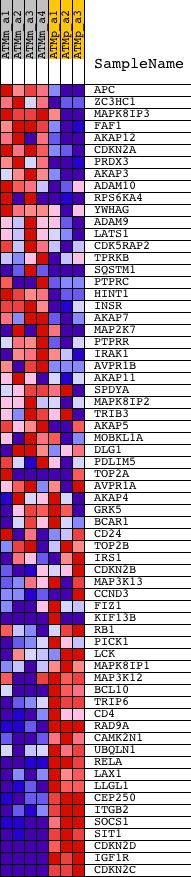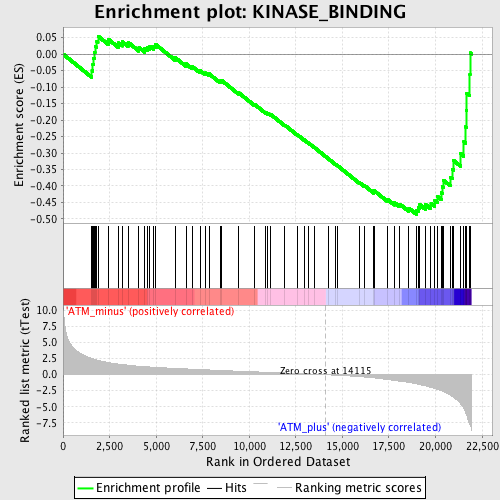
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | KINASE_BINDING |
| Enrichment Score (ES) | -0.48644793 |
| Normalized Enrichment Score (NES) | -1.6777855 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.29951292 |
| FWER p-Value | 0.992 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 1550 | 2.542 | -0.0497 | No | ||
| 2 | ZC3HC1 | 1426654_at | 1597 | 2.485 | -0.0310 | No | ||
| 3 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 1626 | 2.452 | -0.0118 | No | ||
| 4 | FAF1 | 1418242_at 1434370_s_at | 1691 | 2.394 | 0.0052 | No | ||
| 5 | AKAP12 | 1419706_a_at 1458235_at | 1716 | 2.371 | 0.0239 | No | ||
| 6 | CDKN2A | 1450140_a_at | 1811 | 2.297 | 0.0388 | No | ||
| 7 | PRDX3 | 1416292_at | 1874 | 2.238 | 0.0546 | No | ||
| 8 | AKAP3 | 1421531_at 1456591_x_at | 2429 | 1.874 | 0.0449 | No | ||
| 9 | ADAM10 | 1450104_at 1460083_at | 2970 | 1.654 | 0.0340 | No | ||
| 10 | RPS6KA4 | 1438243_at 1448498_at | 3178 | 1.576 | 0.0377 | No | ||
| 11 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 3489 | 1.477 | 0.0358 | No | ||
| 12 | ADAM9 | 1416094_at | 4074 | 1.314 | 0.0201 | No | ||
| 13 | LATS1 | 1427679_at 1460264_at | 4388 | 1.251 | 0.0162 | No | ||
| 14 | CDK5RAP2 | 1443976_at 1447103_at 1452618_at | 4532 | 1.226 | 0.0199 | No | ||
| 15 | TPRKB | 1424992_at 1425410_at 1425682_a_at | 4658 | 1.200 | 0.0242 | No | ||
| 16 | SQSTM1 | 1440076_at 1442237_at 1444021_at 1450957_a_at | 4868 | 1.159 | 0.0243 | No | ||
| 17 | PTPRC | 1422124_a_at 1440165_at | 4964 | 1.140 | 0.0295 | No | ||
| 18 | HINT1 | 1424017_a_at 1424018_at 1457286_at | 6018 | 0.984 | -0.0105 | No | ||
| 19 | INSR | 1421380_at 1450225_at | 6601 | 0.897 | -0.0296 | No | ||
| 20 | AKAP7 | 1422143_at 1433905_at | 6932 | 0.849 | -0.0376 | No | ||
| 21 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 7359 | 0.793 | -0.0504 | No | ||
| 22 | PTPRR | 1426047_a_at | 7642 | 0.757 | -0.0570 | No | ||
| 23 | IRAK1 | 1448668_a_at 1460649_at | 7836 | 0.731 | -0.0597 | No | ||
| 24 | AVPR1B | 1422204_at | 8426 | 0.654 | -0.0812 | No | ||
| 25 | AKAP11 | 1434764_at | 8505 | 0.645 | -0.0794 | No | ||
| 26 | SPDYA | 1429470_at 1432134_at 1441390_at | 9422 | 0.536 | -0.1168 | No | ||
| 27 | MAPK8IP2 | 1418785_at 1418786_at 1435045_s_at 1449225_a_at 1455194_at | 10262 | 0.442 | -0.1515 | No | ||
| 28 | TRIB3 | 1426065_a_at 1456225_x_at | 10879 | 0.375 | -0.1765 | No | ||
| 29 | AKAP5 | 1447301_at | 11001 | 0.361 | -0.1790 | No | ||
| 30 | MOBKL1A | 1430564_at 1455184_at | 11163 | 0.345 | -0.1835 | No | ||
| 31 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 11911 | 0.259 | -0.2155 | No | ||
| 32 | PDLIM5 | 1429783_at 1438186_at 1442710_at 1446339_at | 12596 | 0.189 | -0.2452 | No | ||
| 33 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 12942 | 0.148 | -0.2598 | No | ||
| 34 | AVPR1A | 1418603_at 1418604_at | 13189 | 0.118 | -0.2700 | No | ||
| 35 | AKAP4 | 1419019_a_at 1431868_at 1451661_at 1451662_x_at | 13511 | 0.080 | -0.2840 | No | ||
| 36 | GRK5 | 1446361_at 1446998_at 1449514_at | 14266 | -0.024 | -0.3183 | No | ||
| 37 | BCAR1 | 1439388_s_at 1450622_at | 14615 | -0.077 | -0.3336 | No | ||
| 38 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 14737 | -0.094 | -0.3383 | No | ||
| 39 | TOP2B | 1416731_at 1416732_at 1448458_at 1456688_at | 15901 | -0.301 | -0.3890 | No | ||
| 40 | IRS1 | 1423104_at | 16185 | -0.367 | -0.3989 | No | ||
| 41 | CDKN2B | 1449152_at | 16689 | -0.493 | -0.4177 | No | ||
| 42 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 16696 | -0.498 | -0.4139 | No | ||
| 43 | CCND3 | 1415907_at 1437584_at 1444323_at 1446567_at 1447434_at 1457549_at | 17403 | -0.737 | -0.4400 | No | ||
| 44 | FIZ1 | 1423733_a_at | 17794 | -0.901 | -0.4503 | No | ||
| 45 | KIF13B | 1433158_at 1433159_at 1436056_at 1453276_at | 18077 | -1.006 | -0.4548 | No | ||
| 46 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 18569 | -1.202 | -0.4672 | No | ||
| 47 | PICK1 | 1419384_at | 18990 | -1.425 | -0.4746 | Yes | ||
| 48 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 19071 | -1.481 | -0.4659 | Yes | ||
| 49 | MAPK8IP1 | 1425679_a_at 1440619_at | 19157 | -1.537 | -0.4569 | Yes | ||
| 50 | MAP3K12 | 1417115_at 1438908_at 1456565_s_at | 19465 | -1.743 | -0.4564 | Yes | ||
| 51 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 19756 | -1.997 | -0.4530 | Yes | ||
| 52 | TRIP6 | 1449041_a_at | 19965 | -2.144 | -0.4446 | Yes | ||
| 53 | CD4 | 1419696_at 1427779_a_at | 20116 | -2.283 | -0.4324 | Yes | ||
| 54 | RAD9A | 1418404_at | 20313 | -2.489 | -0.4206 | Yes | ||
| 55 | CAMK2N1 | 1456609_at | 20362 | -2.545 | -0.4016 | Yes | ||
| 56 | UBQLN1 | 1419385_a_at 1424368_s_at 1450345_at | 20418 | -2.623 | -0.3822 | Yes | ||
| 57 | RELA | 1419536_a_at | 20799 | -3.184 | -0.3730 | Yes | ||
| 58 | LAX1 | 1438687_at | 20919 | -3.427 | -0.3499 | Yes | ||
| 59 | LLGL1 | 1416621_at | 20971 | -3.528 | -0.3228 | Yes | ||
| 60 | CEP250 | 1421512_at | 21354 | -4.579 | -0.3021 | Yes | ||
| 61 | ITGB2 | 1450678_at | 21523 | -5.260 | -0.2659 | Yes | ||
| 62 | SOCS1 | 1440047_at 1450446_a_at | 21625 | -5.963 | -0.2207 | Yes | ||
| 63 | SIT1 | 1418751_at | 21653 | -6.137 | -0.1708 | Yes | ||
| 64 | CDKN2D | 1416253_at | 21682 | -6.381 | -0.1188 | Yes | ||
| 65 | IGF1R | 1426565_at 1437219_at 1443566_at 1445600_at 1445664_at 1446303_at | 21847 | -7.797 | -0.0613 | Yes | ||
| 66 | CDKN2C | 1416868_at 1439164_at | 21857 | -7.825 | 0.0036 | Yes |