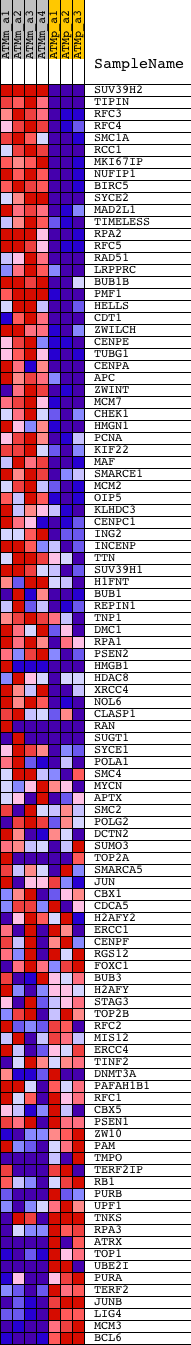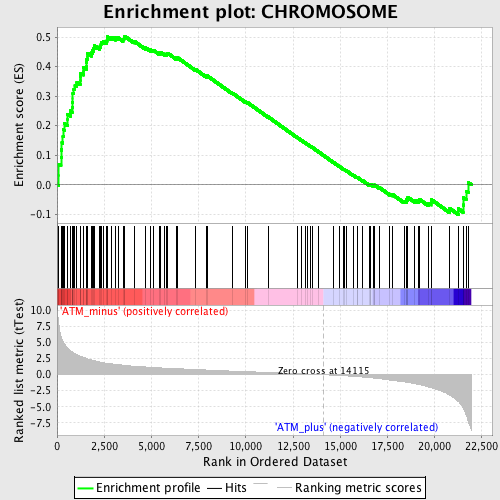
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | CHROMOSOME |
| Enrichment Score (ES) | 0.503004 |
| Normalized Enrichment Score (NES) | 1.8912053 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0142827695 |
| FWER p-Value | 0.188 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SUV39H2 | 1422979_at 1433996_at 1436561_at | 90 | 7.866 | 0.0328 | Yes | ||
| 2 | TIPIN | 1426612_at | 93 | 7.774 | 0.0692 | Yes | ||
| 3 | RFC3 | 1423700_at 1432538_a_at | 206 | 5.897 | 0.0918 | Yes | ||
| 4 | RFC4 | 1424321_at 1438161_s_at | 219 | 5.766 | 0.1183 | Yes | ||
| 5 | SMC1A | 1417830_at 1417831_at 1417832_at | 256 | 5.456 | 0.1423 | Yes | ||
| 6 | RCC1 | 1416962_at | 309 | 5.142 | 0.1641 | Yes | ||
| 7 | MKI67IP | 1424001_at | 334 | 5.045 | 0.1867 | Yes | ||
| 8 | NUFIP1 | 1448802_at | 375 | 4.848 | 0.2076 | Yes | ||
| 9 | BIRC5 | 1424278_a_at | 545 | 4.191 | 0.2196 | Yes | ||
| 10 | SYCE2 | 1429270_a_at | 551 | 4.184 | 0.2390 | Yes | ||
| 11 | MAD2L1 | 1422460_at | 698 | 3.780 | 0.2501 | Yes | ||
| 12 | TIMELESS | 1417586_at 1417587_at | 796 | 3.555 | 0.2623 | Yes | ||
| 13 | RPA2 | 1416433_at 1454011_a_at | 818 | 3.519 | 0.2779 | Yes | ||
| 14 | RFC5 | 1452917_at | 835 | 3.489 | 0.2935 | Yes | ||
| 15 | RAD51 | 1418281_at | 836 | 3.487 | 0.3099 | Yes | ||
| 16 | LRPPRC | 1424353_at | 887 | 3.375 | 0.3235 | Yes | ||
| 17 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 946 | 3.263 | 0.3361 | Yes | ||
| 18 | PMF1 | 1416707_a_at 1438173_x_at | 1048 | 3.105 | 0.3461 | Yes | ||
| 19 | HELLS | 1417541_at 1430139_at 1453361_at | 1219 | 2.886 | 0.3519 | Yes | ||
| 20 | CDT1 | 1424143_a_at 1424144_at 1455791_at | 1225 | 2.881 | 0.3652 | Yes | ||
| 21 | ZWILCH | 1416757_at | 1239 | 2.858 | 0.3780 | Yes | ||
| 22 | CENPE | 1435005_at 1439040_at | 1385 | 2.705 | 0.3841 | Yes | ||
| 23 | TUBG1 | 1417144_at | 1390 | 2.702 | 0.3966 | Yes | ||
| 24 | CENPA | 1441864_x_at 1444416_at 1450842_a_at | 1542 | 2.550 | 0.4016 | Yes | ||
| 25 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 1550 | 2.542 | 0.4133 | Yes | ||
| 26 | ZWINT | 1423724_at 1427539_a_at 1427540_at 1429786_a_at 1429787_x_at 1444717_at 1455382_at | 1573 | 2.514 | 0.4241 | Yes | ||
| 27 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 1610 | 2.476 | 0.4340 | Yes | ||
| 28 | CHEK1 | 1420031_at 1420032_at 1439208_at 1449708_s_at 1450677_at | 1615 | 2.468 | 0.4454 | Yes | ||
| 29 | HMGN1 | 1422495_a_at 1422496_at 1438940_x_at 1455897_x_at | 1834 | 2.273 | 0.4461 | Yes | ||
| 30 | PCNA | 1417947_at | 1885 | 2.232 | 0.4543 | Yes | ||
| 31 | KIF22 | 1423813_at 1437716_x_at 1451128_s_at | 1944 | 2.185 | 0.4619 | Yes | ||
| 32 | MAF | 1435828_at 1437473_at 1444073_at 1447849_s_at 1447945_at 1456060_at | 1970 | 2.170 | 0.4710 | Yes | ||
| 33 | SMARCE1 | 1422675_at 1422676_at 1430822_at | 2269 | 1.970 | 0.4666 | Yes | ||
| 34 | MCM2 | 1434079_s_at 1448777_at | 2322 | 1.939 | 0.4733 | Yes | ||
| 35 | OIP5 | 1430617_at | 2329 | 1.935 | 0.4821 | Yes | ||
| 36 | KLHDC3 | 1415991_a_at 1448164_at 1454747_a_at 1455910_at | 2443 | 1.868 | 0.4857 | Yes | ||
| 37 | CENPC1 | 1420441_at | 2610 | 1.785 | 0.4865 | Yes | ||
| 38 | ING2 | 1419111_at | 2644 | 1.771 | 0.4933 | Yes | ||
| 39 | INCENP | 1423092_at 1423093_at 1439436_x_at 1441314_at | 2653 | 1.768 | 0.5012 | Yes | ||
| 40 | TTN | 1427445_a_at 1427446_s_at 1431928_at 1444083_at 1444638_at 1446450_at | 2890 | 1.686 | 0.4984 | Yes | ||
| 41 | SUV39H1 | 1427382_a_at 1432227_at 1432236_a_at | 3081 | 1.612 | 0.4972 | Yes | ||
| 42 | H1FNT | 1430181_at 1441954_s_at | 3224 | 1.560 | 0.4980 | Yes | ||
| 43 | BUB1 | 1424046_at 1438571_at | 3491 | 1.476 | 0.4928 | Yes | ||
| 44 | REPIN1 | 1425535_at 1434044_at 1457387_at | 3543 | 1.461 | 0.4973 | Yes | ||
| 45 | TNP1 | 1415924_at 1438632_x_at | 3569 | 1.454 | 0.5030 | Yes | ||
| 46 | DMC1 | 1420335_at 1449819_at | 4079 | 1.313 | 0.4859 | No | ||
| 47 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 4687 | 1.195 | 0.4637 | No | ||
| 48 | PSEN2 | 1425869_a_at 1431542_at | 4952 | 1.143 | 0.4569 | No | ||
| 49 | HMGB1 | 1416176_at 1425048_a_at 1435324_x_at | 5118 | 1.113 | 0.4546 | No | ||
| 50 | HDAC8 | 1428933_at | 5446 | 1.056 | 0.4446 | No | ||
| 51 | XRCC4 | 1424601_at 1424602_s_at 1445922_at | 5488 | 1.050 | 0.4477 | No | ||
| 52 | NOL6 | 1424472_at 1425629_a_at | 5705 | 1.023 | 0.4426 | No | ||
| 53 | CLASP1 | 1427353_at 1452265_at 1459233_at 1460102_at | 5816 | 1.007 | 0.4423 | No | ||
| 54 | RAN | 1443131_at 1446819_at 1447478_at 1460551_at | 5842 | 1.004 | 0.4458 | No | ||
| 55 | SUGT1 | 1426424_at 1426425_at 1446757_at | 6339 | 0.936 | 0.4275 | No | ||
| 56 | SYCE1 | 1428685_at | 6390 | 0.929 | 0.4296 | No | ||
| 57 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 7331 | 0.795 | 0.3903 | No | ||
| 58 | SMC4 | 1427275_at 1427276_at 1441677_at 1452197_at | 7924 | 0.721 | 0.3665 | No | ||
| 59 | MYCN | 1417155_at 1425922_a_at 1425923_at | 7938 | 0.720 | 0.3693 | No | ||
| 60 | APTX | 1450921_at | 9289 | 0.553 | 0.3101 | No | ||
| 61 | SMC2 | 1429658_a_at 1429659_at 1429660_s_at 1448635_at 1458479_at | 9972 | 0.473 | 0.2811 | No | ||
| 62 | POLG2 | 1450816_at | 10090 | 0.458 | 0.2778 | No | ||
| 63 | DCTN2 | 1424461_at | 11170 | 0.343 | 0.2300 | No | ||
| 64 | SUMO3 | 1422457_s_at 1439372_at | 12709 | 0.176 | 0.1604 | No | ||
| 65 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 12942 | 0.148 | 0.1505 | No | ||
| 66 | SMARCA5 | 1440048_at | 13154 | 0.123 | 0.1414 | No | ||
| 67 | JUN | 1417409_at 1448694_at | 13265 | 0.110 | 0.1369 | No | ||
| 68 | CBX1 | 1422935_x_at 1431773_at 1433718_a_at 1436266_x_at 1450797_a_at | 13442 | 0.089 | 0.1292 | No | ||
| 69 | CDCA5 | 1416802_a_at 1448466_at | 13512 | 0.080 | 0.1264 | No | ||
| 70 | H2AFY2 | 1426363_x_at 1444008_at 1447241_at | 13864 | 0.031 | 0.1105 | No | ||
| 71 | ERCC1 | 1417328_at 1437447_s_at 1441838_at 1445266_at | 14619 | -0.077 | 0.0763 | No | ||
| 72 | CENPF | 1427161_at 1452334_at 1458447_at | 14933 | -0.127 | 0.0626 | No | ||
| 73 | RGS12 | 1429380_at 1453129_a_at 1457511_at 1459566_at | 15167 | -0.164 | 0.0527 | No | ||
| 74 | FOXC1 | 1419485_at 1419486_at | 15213 | -0.172 | 0.0514 | No | ||
| 75 | BUB3 | 1416815_s_at 1448058_s_at 1448473_at 1459104_at 1459918_at | 15306 | -0.186 | 0.0481 | No | ||
| 76 | H2AFY | 1424572_a_at | 15692 | -0.258 | 0.0317 | No | ||
| 77 | STAG3 | 1460229_at | 15884 | -0.298 | 0.0243 | No | ||
| 78 | TOP2B | 1416731_at 1416732_at 1448458_at 1456688_at | 15901 | -0.301 | 0.0250 | No | ||
| 79 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 16192 | -0.368 | 0.0135 | No | ||
| 80 | MIS12 | 1424717_at | 16561 | -0.461 | -0.0012 | No | ||
| 81 | ERCC4 | 1432272_a_at 1440237_at 1441003_at 1448984_at | 16584 | -0.469 | -0.0000 | No | ||
| 82 | TINF2 | 1437166_at 1451163_at | 16615 | -0.476 | 0.0008 | No | ||
| 83 | DNMT3A | 1423063_at 1423064_at 1423065_at 1423066_at 1442309_at 1460324_at | 16777 | -0.524 | -0.0041 | No | ||
| 84 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 16791 | -0.527 | -0.0022 | No | ||
| 85 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 16812 | -0.533 | -0.0006 | No | ||
| 86 | CBX5 | 1421932_at 1421933_at 1421934_at 1450416_at 1454636_at 1459845_at | 17080 | -0.619 | -0.0099 | No | ||
| 87 | PSEN1 | 1420261_at 1421853_at 1425549_at 1450399_at | 17630 | -0.831 | -0.0312 | No | ||
| 88 | ZW10 | 1424891_a_at 1431747_at 1445206_at | 17780 | -0.894 | -0.0338 | No | ||
| 89 | PAM | 1418908_at 1445797_at | 18380 | -1.112 | -0.0560 | No | ||
| 90 | TMPO | 1421237_at 1426020_at 1426349_s_at 1428976_at 1452036_a_at | 18495 | -1.167 | -0.0558 | No | ||
| 91 | TERF2IP | 1419381_at 1456400_at | 18509 | -1.176 | -0.0508 | No | ||
| 92 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 18569 | -1.202 | -0.0479 | No | ||
| 93 | PURB | 1419640_at 1419641_at 1419642_at 1428254_at 1436940_at | 18577 | -1.204 | -0.0426 | No | ||
| 94 | UPF1 | 1419685_at | 18934 | -1.394 | -0.0523 | No | ||
| 95 | TNKS | 1432630_at 1433778_at 1445358_at 1459910_at | 19138 | -1.525 | -0.0545 | No | ||
| 96 | RPA3 | 1448938_at | 19181 | -1.555 | -0.0491 | No | ||
| 97 | ATRX | 1420945_at 1420946_at 1420947_at 1438750_at 1441014_at 1441771_at 1450051_at 1453734_at 1456832_at | 19667 | -1.929 | -0.0622 | No | ||
| 98 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 19804 | -2.024 | -0.0590 | No | ||
| 99 | UBE2I | 1422712_a_at 1422713_a_at 1425478_x_at 1453189_at | 19812 | -2.030 | -0.0498 | No | ||
| 100 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 20789 | -3.168 | -0.0796 | No | ||
| 101 | TERF2 | 1421147_at 1455651_at 1459650_at | 21248 | -4.163 | -0.0810 | No | ||
| 102 | JUNB | 1415899_at | 21503 | -5.184 | -0.0683 | No | ||
| 103 | LIG4 | 1439487_at | 21518 | -5.251 | -0.0443 | No | ||
| 104 | MCM3 | 1420029_at 1426653_at | 21664 | -6.234 | -0.0216 | No | ||
| 105 | BCL6 | 1421818_at 1450381_a_at | 21780 | -7.247 | 0.0071 | No |