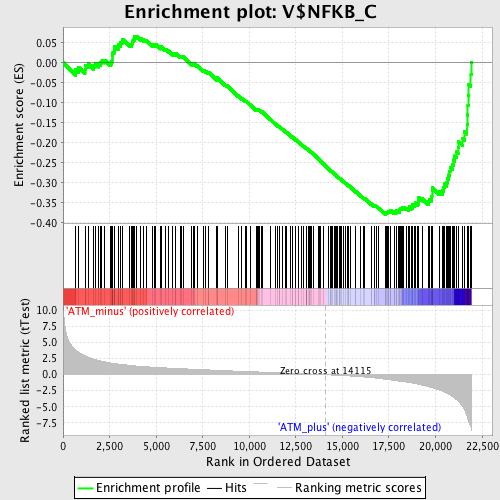
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$NFKB_C |
| Enrichment Score (ES) | -0.3788015 |
| Normalized Enrichment Score (NES) | -1.5474565 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09541468 |
| FWER p-Value | 0.712 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARHGAP8 | 1451463_at | 682 | 3.837 | -0.0171 | No | ||
| 2 | ECT2 | 1419513_a_at | 839 | 3.479 | -0.0113 | No | ||
| 3 | UACA | 1452985_at | 1176 | 2.932 | -0.0158 | No | ||
| 4 | TCEA2 | 1418107_at 1440791_x_at | 1209 | 2.894 | -0.0065 | No | ||
| 5 | PIGC | 1424189_at 1424190_at | 1341 | 2.754 | -0.0022 | No | ||
| 6 | PURG | 1424970_at 1452029_a_at | 1645 | 2.434 | -0.0071 | No | ||
| 7 | CGGBP1 | 1454641_at 1455658_at | 1723 | 2.368 | -0.0018 | No | ||
| 8 | SOCS2 | 1418507_s_at 1438470_at 1441476_at 1442586_at 1446085_at 1449109_at | 1916 | 2.210 | -0.0024 | No | ||
| 9 | UNC84B | 1433832_at 1439238_at | 2002 | 2.144 | 0.0017 | No | ||
| 10 | IL2RA | 1420691_at 1420692_at | 2087 | 2.083 | 0.0056 | No | ||
| 11 | TRIM47 | 1426784_at | 2223 | 1.994 | 0.0068 | No | ||
| 12 | FOXA3 | 1431900_a_at | 2529 | 1.821 | -0.0005 | No | ||
| 13 | FOXD3 | 1422210_at | 2613 | 1.783 | 0.0024 | No | ||
| 14 | SEC14L2 | 1424530_at | 2627 | 1.777 | 0.0084 | No | ||
| 15 | CDC37 | 1416819_at 1416820_at | 2632 | 1.776 | 0.0148 | No | ||
| 16 | FOXD2 | 1450289_at | 2635 | 1.775 | 0.0214 | No | ||
| 17 | GRIN2D | 1421393_at | 2675 | 1.760 | 0.0261 | No | ||
| 18 | CLCN1 | 1427591_at 1448082_at | 2734 | 1.737 | 0.0299 | No | ||
| 19 | HCFC1 | 1421972_s_at 1433736_at 1445688_at 1446868_at 1450439_at 1457875_at | 2753 | 1.731 | 0.0356 | No | ||
| 20 | FGF17 | 1421523_at 1456239_at | 2777 | 1.727 | 0.0409 | No | ||
| 21 | SMPD3 | 1422779_at 1438665_at 1445759_at 1450748_at | 2953 | 1.663 | 0.0391 | No | ||
| 22 | SMARCAD1 | 1435651_a_at 1440066_at 1452276_at | 2963 | 1.659 | 0.0449 | No | ||
| 23 | ENO3 | 1417951_at 1449631_at | 3066 | 1.617 | 0.0462 | No | ||
| 24 | COL11A2 | 1423578_at | 3090 | 1.609 | 0.0511 | No | ||
| 25 | SOX3 | 1435192_at 1450485_at | 3170 | 1.578 | 0.0534 | No | ||
| 26 | GNAO1 | 1421152_a_at 1448031_at 1460383_at | 3203 | 1.566 | 0.0578 | No | ||
| 27 | ALG6 | 1439007_at 1459337_at | 3578 | 1.451 | 0.0460 | No | ||
| 28 | PTGES | 1439747_at 1449449_at 1449450_at | 3686 | 1.412 | 0.0463 | No | ||
| 29 | PTHLH | 1422324_a_at 1427527_a_at | 3701 | 1.409 | 0.0509 | No | ||
| 30 | RAD18 | 1423318_at 1435826_at 1443954_at 1451928_a_at | 3739 | 1.396 | 0.0544 | No | ||
| 31 | CDH5 | 1422047_at 1433956_at | 3777 | 1.387 | 0.0579 | No | ||
| 32 | S100A10 | 1416762_at 1439348_at 1456642_x_at | 3816 | 1.377 | 0.0613 | No | ||
| 33 | PHF6 | 1430415_at 1453761_at 1454625_at | 3824 | 1.376 | 0.0661 | No | ||
| 34 | ASCL3 | 1420780_at | 3925 | 1.352 | 0.0665 | No | ||
| 35 | NDRG2 | 1448154_at | 4161 | 1.295 | 0.0605 | No | ||
| 36 | CTGF | 1416953_at | 4338 | 1.260 | 0.0571 | No | ||
| 37 | HSD3B7 | 1416968_a_at 1442912_at | 4453 | 1.238 | 0.0565 | No | ||
| 38 | BLR1 | 1422003_at | 4777 | 1.176 | 0.0460 | No | ||
| 39 | DNAJA1 | 1416288_at 1437220_x_at 1445729_at 1459835_s_at 1460179_at | 4912 | 1.152 | 0.0442 | No | ||
| 40 | SCRT2 | 1440930_a_at 1440931_at 1446986_at 1457310_x_at | 4977 | 1.136 | 0.0454 | No | ||
| 41 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 5220 | 1.096 | 0.0384 | No | ||
| 42 | CCDC8 | 1444761_at | 5273 | 1.087 | 0.0401 | No | ||
| 43 | HOXB6 | 1451660_a_at | 5486 | 1.050 | 0.0342 | No | ||
| 44 | FUT7 | 1420756_at | 5671 | 1.026 | 0.0296 | No | ||
| 45 | SLC6A12 | 1449382_at | 5898 | 0.999 | 0.0229 | No | ||
| 46 | BCKDK | 1443813_x_at 1460644_at | 6010 | 0.985 | 0.0215 | No | ||
| 47 | HOXA11 | 1420414_at | 6043 | 0.981 | 0.0237 | No | ||
| 48 | GATA4 | 1418863_at 1418864_at 1441364_at | 6287 | 0.944 | 0.0160 | No | ||
| 49 | WRN | 1425074_at 1425982_a_at 1450163_a_at | 6346 | 0.935 | 0.0168 | No | ||
| 50 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 6447 | 0.919 | 0.0156 | No | ||
| 51 | CHX10 | 1419628_at | 6912 | 0.852 | -0.0025 | No | ||
| 52 | AGC1 | 1449827_at | 6990 | 0.841 | -0.0029 | No | ||
| 53 | LAMB3 | 1417812_a_at | 7042 | 0.834 | -0.0022 | No | ||
| 54 | MOBKL2C | 1440919_at 1459919_a_at | 7224 | 0.809 | -0.0075 | No | ||
| 55 | BAI2 | 1435558_at | 7537 | 0.770 | -0.0190 | No | ||
| 56 | C1QL1 | 1422777_at | 7651 | 0.755 | -0.0213 | No | ||
| 57 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 7784 | 0.739 | -0.0247 | No | ||
| 58 | LIN28 | 1437752_at | 7823 | 0.734 | -0.0237 | No | ||
| 59 | HOXB9 | 1452317_at | 8253 | 0.676 | -0.0409 | No | ||
| 60 | CUEDC1 | 1425920_at 1435358_at 1443455_at 1451447_at | 8268 | 0.674 | -0.0390 | No | ||
| 61 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 8288 | 0.671 | -0.0374 | No | ||
| 62 | ANP32A | 1421918_at 1434555_at 1450407_a_at | 8737 | 0.617 | -0.0557 | No | ||
| 63 | LRRFIP2 | 1436378_at 1436510_a_at | 8822 | 0.607 | -0.0573 | No | ||
| 64 | IL13 | 1420802_at | 9410 | 0.537 | -0.0823 | No | ||
| 65 | TNFSF7 | 1449926_at | 9600 | 0.515 | -0.0891 | No | ||
| 66 | ANKHD1 | 1419867_a_at 1436597_at 1440754_at 1441597_at 1442641_at 1448008_at 1453023_at 1455759_a_at | 9779 | 0.496 | -0.0954 | No | ||
| 67 | KCNQ5 | 1427426_at 1441071_at | 9835 | 0.489 | -0.0961 | No | ||
| 68 | WNT10A | 1460657_at | 10065 | 0.462 | -0.1049 | No | ||
| 69 | CYLD | 1429617_at 1429618_at 1445213_at | 10365 | 0.433 | -0.1171 | No | ||
| 70 | CENTB5 | 1439021_at | 10369 | 0.432 | -0.1156 | No | ||
| 71 | BHLHB2 | 1418025_at | 10423 | 0.425 | -0.1165 | No | ||
| 72 | NFKBIB | 1421266_s_at 1446718_at | 10476 | 0.420 | -0.1173 | No | ||
| 73 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 10545 | 0.414 | -0.1189 | No | ||
| 74 | VNN3 | 1420723_at | 10679 | 0.399 | -0.1235 | No | ||
| 75 | LASP1 | 1438633_x_at 1438634_x_at 1439264_x_at 1444947_at 1448207_at 1455470_x_at 1456309_x_at 1456469_x_at 1456578_x_at 1460173_at | 10691 | 0.397 | -0.1225 | No | ||
| 76 | TSNAXIP1 | 1427018_at | 11115 | 0.349 | -0.1407 | No | ||
| 77 | SOX10 | 1424985_a_at 1425369_a_at 1451689_a_at | 11397 | 0.316 | -0.1524 | No | ||
| 78 | EGF | 1418093_a_at | 11531 | 0.301 | -0.1574 | No | ||
| 79 | PCSK2 | 1428305_at 1444147_at 1447991_at 1447992_s_at 1448312_at | 11606 | 0.294 | -0.1598 | No | ||
| 80 | TSLP | 1450004_at | 11613 | 0.293 | -0.1589 | No | ||
| 81 | RGL1 | 1418535_at 1440662_at 1449124_at | 11802 | 0.271 | -0.1666 | No | ||
| 82 | EDG5 | 1428176_at 1447880_x_at | 11941 | 0.257 | -0.1720 | No | ||
| 83 | MSC | 1418417_at | 11985 | 0.252 | -0.1730 | No | ||
| 84 | SYMPK | 1428673_at | 12203 | 0.231 | -0.1821 | No | ||
| 85 | JAK3 | 1425750_a_at 1446515_at | 12294 | 0.222 | -0.1854 | No | ||
| 86 | PLA1A | 1417785_at | 12305 | 0.221 | -0.1851 | No | ||
| 87 | IL23A | 1419529_at | 12481 | 0.201 | -0.1924 | No | ||
| 88 | LCAT | 1417043_at | 12486 | 0.200 | -0.1918 | No | ||
| 89 | RND1 | 1455197_at | 12644 | 0.183 | -0.1984 | No | ||
| 90 | TAL1 | 1449389_at | 12666 | 0.181 | -0.1986 | No | ||
| 91 | IL1RN | 1423017_a_at 1425663_at 1451798_at | 12787 | 0.166 | -0.2035 | No | ||
| 92 | DCAMKL1 | 1423125_at 1424270_at 1424271_at 1436659_at 1446190_at 1450863_a_at 1451289_at 1451917_a_at | 12924 | 0.150 | -0.2092 | No | ||
| 93 | VCAM1 | 1415989_at 1436003_at 1448162_at 1451314_a_at | 13049 | 0.135 | -0.2144 | No | ||
| 94 | AMOTL1 | 1428785_at 1431121_at 1446909_at 1455247_at | 13058 | 0.134 | -0.2143 | No | ||
| 95 | DDR1 | 1415797_at 1415798_at 1435820_x_at 1437619_x_at 1438367_x_at 1439382_x_at 1456226_x_at 1459990_at | 13074 | 0.132 | -0.2145 | No | ||
| 96 | RALGDS | 1460634_at | 13078 | 0.131 | -0.2142 | No | ||
| 97 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 13156 | 0.123 | -0.2172 | No | ||
| 98 | GNG4 | 1417943_at 1417944_at 1447669_s_at | 13231 | 0.115 | -0.2202 | No | ||
| 99 | SLC34A3 | 1439519_at | 13246 | 0.112 | -0.2204 | No | ||
| 100 | PAPPA | 1427633_a_at 1432591_at 1432592_at 1459375_at | 13287 | 0.107 | -0.2219 | No | ||
| 101 | SESN2 | 1425139_at 1451599_at | 13330 | 0.101 | -0.2234 | No | ||
| 102 | PTGS2 | 1417262_at 1417263_at | 13435 | 0.089 | -0.2279 | No | ||
| 103 | LCN2 | 1427747_a_at | 13699 | 0.054 | -0.2398 | No | ||
| 104 | MSX1 | 1417127_at 1448601_s_at | 13774 | 0.044 | -0.2430 | No | ||
| 105 | RIN2 | 1426368_at 1441209_at 1442829_at | 13830 | 0.037 | -0.2454 | No | ||
| 106 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 13996 | 0.016 | -0.2530 | No | ||
| 107 | GRK5 | 1446361_at 1446998_at 1449514_at | 14266 | -0.024 | -0.2652 | No | ||
| 108 | IL1RAPL1 | 1445821_at 1458964_at | 14354 | -0.036 | -0.2691 | No | ||
| 109 | BDNF | 1422168_a_at 1422169_a_at | 14372 | -0.039 | -0.2698 | No | ||
| 110 | DAP3 | 1450848_at | 14419 | -0.045 | -0.2717 | No | ||
| 111 | OPCML | 1435612_at 1442752_at 1457446_at 1458228_at | 14441 | -0.050 | -0.2725 | No | ||
| 112 | PPP2R5E | 1428461_at 1428462_at 1428463_a_at 1439401_x_at 1443533_at 1443949_at 1446356_at 1452788_at | 14554 | -0.068 | -0.2774 | No | ||
| 113 | IL1A | 1421473_at | 14640 | -0.081 | -0.2810 | No | ||
| 114 | MAG | 1460219_at | 14655 | -0.083 | -0.2813 | No | ||
| 115 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 14691 | -0.088 | -0.2826 | No | ||
| 116 | SIX5 | 1427560_at | 14750 | -0.096 | -0.2849 | No | ||
| 117 | MAML2 | 1446570_at 1447546_s_at | 14832 | -0.110 | -0.2882 | No | ||
| 118 | SNAP25 | 1416828_at 1443398_at | 14853 | -0.114 | -0.2887 | No | ||
| 119 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 14894 | -0.121 | -0.2901 | No | ||
| 120 | GABPB2 | 1440289_at 1440868_at 1443942_at 1453682_at 1455110_at | 14930 | -0.126 | -0.2913 | No | ||
| 121 | PLP1 | 1425467_a_at 1425468_at 1451718_at | 15053 | -0.146 | -0.2963 | No | ||
| 122 | ITGB4 | 1427387_a_at | 15187 | -0.168 | -0.3018 | No | ||
| 123 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 15251 | -0.177 | -0.3041 | No | ||
| 124 | TYRO3 | 1425248_a_at 1425249_a_at | 15300 | -0.185 | -0.3056 | No | ||
| 125 | PTPRJ | 1425587_a_at 1425588_at 1427602_at 1427629_at 1443683_at 1455030_at | 15352 | -0.195 | -0.3072 | No | ||
| 126 | ZADH2 | 1451277_at | 15439 | -0.210 | -0.3104 | No | ||
| 127 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 15724 | -0.265 | -0.3224 | No | ||
| 128 | ITGB8 | 1436223_at | 15727 | -0.265 | -0.3215 | No | ||
| 129 | LYPLA2 | 1417433_at | 15994 | -0.322 | -0.3326 | No | ||
| 130 | EFNA3 | 1431170_at 1455344_at | 16158 | -0.359 | -0.3387 | No | ||
| 131 | CXCL10 | 1418930_at | 16198 | -0.369 | -0.3392 | No | ||
| 132 | IFNB1 | 1422305_at | 16546 | -0.456 | -0.3534 | No | ||
| 133 | LRCH1 | 1438032_at 1442502_at 1442757_at 1445396_at 1447746_at 1459150_at | 16558 | -0.460 | -0.3522 | No | ||
| 134 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 16696 | -0.498 | -0.3567 | No | ||
| 135 | DACH2 | 1449823_at 1458012_at | 16744 | -0.513 | -0.3569 | No | ||
| 136 | JPH3 | 1418161_at 1443724_at | 16805 | -0.530 | -0.3577 | No | ||
| 137 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 16963 | -0.583 | -0.3627 | No | ||
| 138 | IL18R1 | 1421628_at | 17313 | -0.698 | -0.3762 | Yes | ||
| 139 | RHOA | 1437628_s_at | 17319 | -0.701 | -0.3738 | Yes | ||
| 140 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 17391 | -0.733 | -0.3744 | Yes | ||
| 141 | IER5 | 1417612_at 1417613_at | 17447 | -0.757 | -0.3741 | Yes | ||
| 142 | CD74 | 1425519_a_at | 17456 | -0.760 | -0.3716 | Yes | ||
| 143 | ID2 | 1422537_a_at 1435176_a_at 1453596_at | 17499 | -0.777 | -0.3706 | Yes | ||
| 144 | ASH1L | 1420985_at 1441117_at 1450071_at 1450072_at | 17568 | -0.804 | -0.3708 | Yes | ||
| 145 | RAP2C | 1428119_a_at 1437016_x_at 1460430_at | 17572 | -0.806 | -0.3679 | Yes | ||
| 146 | FGF1 | 1423136_at 1441042_at 1450869_at | 17775 | -0.893 | -0.3739 | Yes | ||
| 147 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 17819 | -0.912 | -0.3724 | Yes | ||
| 148 | FOXJ2 | 1420373_at 1420374_at | 17880 | -0.936 | -0.3717 | Yes | ||
| 149 | PPP3CA | 1426401_at 1438478_a_at 1440051_at 1452056_s_at 1458495_at | 17912 | -0.953 | -0.3696 | Yes | ||
| 150 | TCTA | 1420123_at 1420124_s_at 1420125_at 1424068_at | 17987 | -0.975 | -0.3693 | Yes | ||
| 151 | GPHN | 1426462_at 1426463_at 1430038_at 1444869_at 1445188_at 1458242_at | 18074 | -1.005 | -0.3696 | Yes | ||
| 152 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 18087 | -1.008 | -0.3663 | Yes | ||
| 153 | GNA13 | 1422555_s_at 1422556_at 1430295_at 1433749_at 1446722_at 1450656_at 1453470_a_at 1460317_s_at | 18114 | -1.016 | -0.3638 | Yes | ||
| 154 | ARPC5 | 1444568_at 1448129_at | 18190 | -1.036 | -0.3633 | Yes | ||
| 155 | BMF | 1422995_at 1454880_s_at 1458325_x_at | 18237 | -1.053 | -0.3615 | Yes | ||
| 156 | PPP1R13B | 1434895_s_at 1442956_at 1459224_at | 18299 | -1.081 | -0.3603 | Yes | ||
| 157 | SIN3A | 1419101_at 1419102_at 1419900_at 1449343_s_at | 18444 | -1.142 | -0.3627 | Yes | ||
| 158 | NFKB2 | 1425902_a_at 1429128_x_at | 18571 | -1.202 | -0.3640 | Yes | ||
| 159 | IL4I1 | 1419192_at | 18576 | -1.204 | -0.3597 | Yes | ||
| 160 | MAP3K8 | 1419208_at | 18698 | -1.260 | -0.3606 | Yes | ||
| 161 | PFN1 | 1449018_at | 18754 | -1.291 | -0.3583 | Yes | ||
| 162 | ICAM1 | 1424067_at | 18780 | -1.313 | -0.3545 | Yes | ||
| 163 | SLAMF8 | 1425294_at | 18870 | -1.357 | -0.3536 | Yes | ||
| 164 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 18903 | -1.376 | -0.3499 | Yes | ||
| 165 | DUSP22 | 1441723_at 1448985_at | 19053 | -1.465 | -0.3513 | Yes | ||
| 166 | EIF5A | 1437859_x_at 1451470_s_at | 19093 | -1.496 | -0.3475 | Yes | ||
| 167 | RHOG | 1422572_at | 19097 | -1.498 | -0.3421 | Yes | ||
| 168 | SIRT2 | 1423507_a_at | 19099 | -1.499 | -0.3365 | Yes | ||
| 169 | MNT | 1418192_at | 19289 | -1.619 | -0.3392 | Yes | ||
| 170 | SEMA3B | 1416644_a_at 1431795_a_at 1448415_a_at | 19610 | -1.885 | -0.3469 | Yes | ||
| 171 | XPO6 | 1422759_a_at 1440492_at 1443737_at | 19650 | -1.913 | -0.3415 | Yes | ||
| 172 | RANBP10 | 1424584_a_at 1424585_at 1446446_at | 19783 | -2.009 | -0.3401 | Yes | ||
| 173 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 19804 | -2.024 | -0.3335 | Yes | ||
| 174 | UBE2I | 1422712_a_at 1422713_a_at 1425478_x_at 1453189_at | 19812 | -2.030 | -0.3263 | Yes | ||
| 175 | IRF1 | 1448436_a_at | 19813 | -2.030 | -0.3187 | Yes | ||
| 176 | BAHD1 | 1433703_s_at | 19824 | -2.039 | -0.3115 | Yes | ||
| 177 | NFAT5 | 1426029_a_at 1438999_a_at 1439328_at 1439805_at 1448966_a_at 1451921_a_at 1458213_at | 20219 | -2.391 | -0.3208 | Yes | ||
| 178 | BCL3 | 1418133_at | 20382 | -2.577 | -0.3186 | Yes | ||
| 179 | RAB10 | 1422664_at 1429296_at 1442463_at 1453095_at | 20450 | -2.653 | -0.3118 | Yes | ||
| 180 | STAT6 | 1421708_a_at 1426353_at | 20465 | -2.672 | -0.3025 | Yes | ||
| 181 | SNX15 | 1424337_at 1456649_at 1458021_at | 20609 | -2.879 | -0.2983 | Yes | ||
| 182 | NUP153 | 1441689_at 1441733_s_at 1452176_at | 20656 | -2.957 | -0.2894 | Yes | ||
| 183 | CCL22 | 1417925_at | 20700 | -3.021 | -0.2801 | Yes | ||
| 184 | TAZ | 1416207_at 1453785_at | 20760 | -3.112 | -0.2712 | Yes | ||
| 185 | MRPS6 | 1424440_at 1447585_s_at | 20783 | -3.159 | -0.2605 | Yes | ||
| 186 | ZHX2 | 1455210_at | 20896 | -3.382 | -0.2530 | Yes | ||
| 187 | FOS | 1423100_at | 20955 | -3.496 | -0.2427 | Yes | ||
| 188 | LTB | 1419135_at | 21037 | -3.669 | -0.2327 | Yes | ||
| 189 | SLC43A1 | 1440007_at 1453255_at | 21129 | -3.865 | -0.2225 | Yes | ||
| 190 | IL27RA | 1449508_at | 21212 | -4.062 | -0.2111 | Yes | ||
| 191 | USP52 | 1426700_a_at | 21235 | -4.128 | -0.1967 | Yes | ||
| 192 | TNIP1 | 1427689_a_at | 21471 | -5.027 | -0.1888 | Yes | ||
| 193 | CCL5 | 1418126_at | 21570 | -5.584 | -0.1725 | Yes | ||
| 194 | EPHB6 | 1418051_at | 21691 | -6.444 | -0.1540 | Yes | ||
| 195 | SMOC1 | 1448321_at | 21703 | -6.526 | -0.1302 | Yes | ||
| 196 | PRKCD | 1422847_a_at 1442256_at | 21741 | -6.917 | -0.1061 | Yes | ||
| 197 | FLI1 | 1422024_at 1433512_at 1441584_at | 21767 | -7.169 | -0.0805 | Yes | ||
| 198 | RORC | 1425792_a_at 1425793_a_at | 21785 | -7.272 | -0.0542 | Yes | ||
| 199 | EGR2 | 1427682_a_at 1427683_at | 21904 | -8.113 | -0.0294 | Yes | ||
| 200 | EGR3 | 1421486_at 1436329_at | 21913 | -8.258 | 0.0011 | Yes |