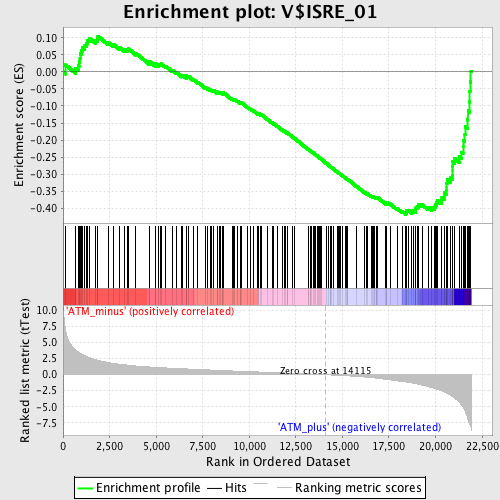
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$ISRE_01 |
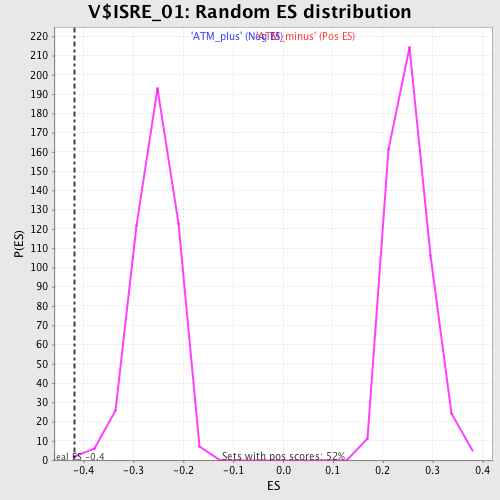
| Enrichment Score (ES) | -0.41858706 |
| Normalized Enrichment Score (NES) | -1.6320971 |
| Nominal p-value | 0.0041753654 |
| FDR q-value | 0.08755092 |
| FWER p-Value | 0.377 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EYA4 | 1432461_at 1442542_at 1445809_at 1456647_a_at 1456665_at | 136 | 6.857 | 0.0205 | No | ||
| 2 | BST2 | 1424921_at | 667 | 3.863 | 0.0113 | No | ||
| 3 | EHD4 | 1449852_a_at | 815 | 3.525 | 0.0183 | No | ||
| 4 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 857 | 3.452 | 0.0299 | No | ||
| 5 | ZFPM1 | 1423603_at 1451046_at | 905 | 3.331 | 0.0407 | No | ||
| 6 | PSMA5 | 1424681_a_at 1434356_a_at | 919 | 3.308 | 0.0530 | No | ||
| 7 | E2F3 | 1427462_at 1434564_at | 989 | 3.187 | 0.0623 | No | ||
| 8 | JARID2 | 1422698_s_at 1446082_at 1450710_at 1456661_at | 1041 | 3.109 | 0.0721 | No | ||
| 9 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 1170 | 2.938 | 0.0777 | No | ||
| 10 | ANGPTL4 | 1417130_s_at 1453410_at | 1264 | 2.835 | 0.0845 | No | ||
| 11 | TCIRG1 | 1420635_a_at | 1322 | 2.769 | 0.0927 | No | ||
| 12 | EPSTI1 | 1432235_at 1444103_at 1452087_at 1454169_a_at | 1424 | 2.671 | 0.0985 | No | ||
| 13 | CD151 | 1420921_at 1424093_x_at 1427525_at 1437670_x_at 1440507_at 1451232_at 1456085_x_at | 1747 | 2.346 | 0.0928 | No | ||
| 14 | PSMA3 | 1448442_a_at | 1853 | 2.256 | 0.0968 | No | ||
| 15 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 1865 | 2.245 | 0.1051 | No | ||
| 16 | GPR65 | 1449175_at | 2427 | 1.874 | 0.0866 | No | ||
| 17 | NPR3 | 1435184_at 1448024_at 1450286_at | 2690 | 1.753 | 0.0815 | No | ||
| 18 | SLC11A2 | 1417584_at 1426441_at 1441709_at 1447333_at 1452078_a_at | 3035 | 1.630 | 0.0720 | No | ||
| 19 | NT5C3 | 1431306_at 1451050_at | 3313 | 1.537 | 0.0653 | No | ||
| 20 | ASPA | 1418472_at | 3445 | 1.495 | 0.0651 | No | ||
| 21 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 3489 | 1.477 | 0.0689 | No | ||
| 22 | SLA | 1420818_at 1420819_at 1441761_at 1447813_x_at | 3913 | 1.354 | 0.0548 | No | ||
| 23 | AKAP6 | 1440859_at 1444357_at | 4649 | 1.201 | 0.0257 | No | ||
| 24 | PITX3 | 1449917_at | 4660 | 1.200 | 0.0299 | No | ||
| 25 | OPTN | 1429778_at 1435679_at | 4974 | 1.137 | 0.0200 | No | ||
| 26 | SCRT2 | 1440930_a_at 1440931_at 1446986_at 1457310_x_at | 4977 | 1.136 | 0.0243 | No | ||
| 27 | TCF15 | 1449592_at | 5145 | 1.108 | 0.0210 | No | ||
| 28 | TNFSF13B | 1445251_at 1458047_at 1460255_at | 5207 | 1.097 | 0.0225 | No | ||
| 29 | USF1 | 1426164_a_at 1448805_at 1459758_at | 5269 | 1.087 | 0.0239 | No | ||
| 30 | GPSN2 | 1416352_s_at 1429681_a_at | 5519 | 1.047 | 0.0165 | No | ||
| 31 | MS4A1 | 1423226_at 1450912_at | 5881 | 1.001 | 0.0039 | No | ||
| 32 | ANAPC1 | 1415680_at 1417570_at 1444271_at | 6084 | 0.974 | -0.0016 | No | ||
| 33 | MIA2 | 1451964_at | 6376 | 0.930 | -0.0113 | No | ||
| 34 | WNT8B | 1421439_at | 6413 | 0.925 | -0.0094 | No | ||
| 35 | LY86 | 1422903_at | 6609 | 0.896 | -0.0148 | No | ||
| 36 | BLNK | 1451780_at | 6620 | 0.895 | -0.0118 | No | ||
| 37 | KRTAP8-1 | 1427211_at | 6728 | 0.881 | -0.0133 | No | ||
| 38 | TAPBP | 1421812_at 1450378_at | 6994 | 0.840 | -0.0222 | No | ||
| 39 | TNFRSF17 | 1420782_at | 7242 | 0.806 | -0.0304 | No | ||
| 40 | PIGR | 1450060_at | 7636 | 0.758 | -0.0455 | No | ||
| 41 | FIGN | 1450268_at | 7779 | 0.740 | -0.0491 | No | ||
| 42 | SEMA6D | 1446536_at 1448060_at 1453055_at 1459318_at | 7922 | 0.721 | -0.0528 | No | ||
| 43 | TLR7 | 1419848_x_at 1422010_at 1449640_at | 7994 | 0.711 | -0.0533 | No | ||
| 44 | MET | 1422990_at | 8068 | 0.700 | -0.0539 | No | ||
| 45 | ERG | 1425370_a_at 1440244_at | 8274 | 0.673 | -0.0607 | No | ||
| 46 | PCDH7 | 1427592_at 1437442_at 1449249_at 1456214_at | 8291 | 0.671 | -0.0588 | No | ||
| 47 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 8419 | 0.655 | -0.0621 | No | ||
| 48 | ZCCHC5 | 1437355_at | 8427 | 0.654 | -0.0599 | No | ||
| 49 | SLC24A1 | 1451647_at | 8577 | 0.637 | -0.0642 | No | ||
| 50 | ERBB2 | 1424919_at | 8578 | 0.636 | -0.0617 | No | ||
| 51 | CDH3 | 1426673_at 1441255_at | 8604 | 0.632 | -0.0604 | No | ||
| 52 | ARHGEF6 | 1429012_at 1442292_at | 9073 | 0.578 | -0.0797 | No | ||
| 53 | EREG | 1419431_at | 9152 | 0.568 | -0.0810 | No | ||
| 54 | CXXC4 | 1437351_at 1438304_at 1456811_at | 9185 | 0.563 | -0.0803 | No | ||
| 55 | PROK2 | 1419681_a_at 1426176_a_at 1451952_at | 9348 | 0.545 | -0.0856 | No | ||
| 56 | HTR2C | 1435513_at 1450477_at | 9506 | 0.527 | -0.0908 | No | ||
| 57 | TBX21 | 1449361_at | 9537 | 0.523 | -0.0901 | No | ||
| 58 | SIX1 | 1427277_at | 9605 | 0.514 | -0.0912 | No | ||
| 59 | LMO1 | 1418478_at | 9907 | 0.480 | -0.1031 | No | ||
| 60 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 10072 | 0.461 | -0.1089 | No | ||
| 61 | CACNA2D2 | 1426185_at | 10233 | 0.445 | -0.1145 | No | ||
| 62 | GSDMDC1 | 1428767_at | 10450 | 0.422 | -0.1227 | No | ||
| 63 | MOV10 | 1416380_at 1447376_at | 10466 | 0.421 | -0.1218 | No | ||
| 64 | IL4 | 1449864_at | 10496 | 0.418 | -0.1215 | No | ||
| 65 | HEPH | 1448696_at | 10614 | 0.406 | -0.1253 | No | ||
| 66 | OSM | 1438767_at | 10664 | 0.401 | -0.1260 | No | ||
| 67 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 10980 | 0.363 | -0.1390 | No | ||
| 68 | KCNE4 | 1418156_at | 11233 | 0.336 | -0.1493 | No | ||
| 69 | RIPK2 | 1421236_at 1450173_at | 11319 | 0.326 | -0.1519 | No | ||
| 70 | BTAF1 | 1435248_a_at 1435249_at 1435953_at | 11529 | 0.302 | -0.1603 | No | ||
| 71 | SDPR | 1416778_at 1416779_at 1443832_s_at | 11774 | 0.274 | -0.1705 | No | ||
| 72 | AMMECR1 | 1430697_at 1450546_at | 11862 | 0.264 | -0.1734 | No | ||
| 73 | SLC25A28 | 1424776_a_at | 11948 | 0.256 | -0.1763 | No | ||
| 74 | ADAM15 | 1416080_at 1425170_a_at 1438760_x_at 1454206_a_at | 12041 | 0.247 | -0.1796 | No | ||
| 75 | GPX1 | 1460671_at | 12051 | 0.246 | -0.1790 | No | ||
| 76 | IFIT2 | 1418293_at | 12303 | 0.221 | -0.1897 | No | ||
| 77 | AGXT2 | 1435838_at | 12420 | 0.208 | -0.1942 | No | ||
| 78 | COL12A1 | 1427391_a_at 1445751_at | 13167 | 0.121 | -0.2280 | No | ||
| 79 | ZFHX1B | 1422748_at 1434298_at 1442393_at 1445518_at 1454200_at 1456389_at | 13171 | 0.121 | -0.2277 | No | ||
| 80 | PPARGC1A | 1434099_at 1434100_x_at 1437751_at 1456395_at 1460336_at | 13278 | 0.108 | -0.2321 | No | ||
| 81 | LRP2 | 1427133_s_at 1427621_at 1445901_at 1452320_at | 13305 | 0.105 | -0.2329 | No | ||
| 82 | EGFL6 | 1419332_at | 13351 | 0.099 | -0.2346 | No | ||
| 83 | PLXNC1 | 1423213_at 1423214_at 1443433_at 1443434_s_at 1450905_at 1450906_at | 13432 | 0.090 | -0.2379 | No | ||
| 84 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 13474 | 0.085 | -0.2395 | No | ||
| 85 | CXXC5 | 1431469_a_at 1448960_at | 13566 | 0.073 | -0.2434 | No | ||
| 86 | KYNU | 1430570_at 1451903_at | 13683 | 0.055 | -0.2485 | No | ||
| 87 | THRAP1 | 1436904_at 1438725_at 1444068_at 1446840_at 1453160_at 1453161_at | 13689 | 0.055 | -0.2485 | No | ||
| 88 | MSX1 | 1417127_at 1448601_s_at | 13774 | 0.044 | -0.2522 | No | ||
| 89 | HOMER1 | 1421768_a_at 1425671_at 1425710_a_at 1437363_at 1439662_at | 13821 | 0.038 | -0.2542 | No | ||
| 90 | B2M | 1427511_at 1449289_a_at 1452428_a_at | 13859 | 0.032 | -0.2557 | No | ||
| 91 | ESR1 | 1421244_at 1435663_at 1457877_at 1460591_at | 14124 | -0.002 | -0.2679 | No | ||
| 92 | CHKA | 1442277_at 1445795_at 1450264_a_at 1453582_at | 14235 | -0.019 | -0.2728 | No | ||
| 93 | IL1RAPL1 | 1445821_at 1458964_at | 14354 | -0.036 | -0.2781 | No | ||
| 94 | BDNF | 1422168_a_at 1422169_a_at | 14372 | -0.039 | -0.2787 | No | ||
| 95 | CXCL11 | 1419697_at 1419698_at | 14439 | -0.049 | -0.2816 | No | ||
| 96 | DDX58 | 1436562_at 1446457_at 1456890_at | 14531 | -0.064 | -0.2855 | No | ||
| 97 | KCNN3 | 1421632_at 1459308_at | 14722 | -0.092 | -0.2939 | No | ||
| 98 | RRBP1 | 1426123_a_at 1436660_at 1449221_a_at 1452767_at | 14785 | -0.102 | -0.2963 | No | ||
| 99 | FGF9 | 1420795_at 1438718_at 1445258_at | 14829 | -0.109 | -0.2979 | No | ||
| 100 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 14894 | -0.121 | -0.3003 | No | ||
| 101 | KLF14 | 1457397_at | 14987 | -0.135 | -0.3040 | No | ||
| 102 | GATA6 | 1425463_at 1425464_at 1443081_at | 15142 | -0.160 | -0.3105 | No | ||
| 103 | NKX2-2 | 1421112_at | 15245 | -0.177 | -0.3145 | No | ||
| 104 | KPNA3 | 1421828_at 1445527_at 1446538_at 1450385_at 1450386_at | 15286 | -0.183 | -0.3156 | No | ||
| 105 | UNC5C | 1419592_at 1439174_at 1440698_at 1449522_at | 15296 | -0.185 | -0.3153 | No | ||
| 106 | OTX1 | 1437601_at | 15758 | -0.273 | -0.3354 | No | ||
| 107 | CXCL10 | 1418930_at | 16198 | -0.369 | -0.3541 | No | ||
| 108 | TNRC4 | 1452851_at | 16303 | -0.397 | -0.3574 | No | ||
| 109 | OVOL1 | 1419051_at 1419052_at | 16334 | -0.405 | -0.3572 | No | ||
| 110 | IFNB1 | 1422305_at | 16546 | -0.456 | -0.3651 | No | ||
| 111 | USP44 | 1456690_at | 16631 | -0.480 | -0.3671 | No | ||
| 112 | IRF2 | 1418265_s_at 1447527_at | 16673 | -0.489 | -0.3670 | No | ||
| 113 | PRDM16 | 1429308_at 1429309_at 1440745_at 1440870_at 1443251_at | 16738 | -0.510 | -0.3680 | No | ||
| 114 | PRKACA | 1447720_x_at 1450519_a_at | 16824 | -0.536 | -0.3698 | No | ||
| 115 | ZF | 1425698_a_at 1438302_at 1452856_at 1452857_at | 16834 | -0.539 | -0.3681 | No | ||
| 116 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 16884 | -0.556 | -0.3682 | No | ||
| 117 | KCNJ16 | 1435094_at 1436163_at | 17332 | -0.705 | -0.3860 | No | ||
| 118 | HDAC4 | 1436758_at 1447566_at 1454693_at | 17390 | -0.733 | -0.3857 | No | ||
| 119 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 17391 | -0.733 | -0.3828 | No | ||
| 120 | AIF1 | 1418204_s_at | 17564 | -0.804 | -0.3876 | No | ||
| 121 | MAP2K6 | 1426850_a_at 1459292_at | 17943 | -0.963 | -0.4012 | No | ||
| 122 | ADD3 | 1423297_at 1423298_at 1426574_a_at 1447077_at 1456162_x_at | 18215 | -1.045 | -0.4096 | No | ||
| 123 | LGP2 | 1420768_a_at 1420769_at 1451426_at | 18412 | -1.127 | -0.4142 | Yes | ||
| 124 | GNB4 | 1419469_at 1419470_at | 18442 | -1.141 | -0.4111 | Yes | ||
| 125 | HIST1H1C | 1416101_a_at 1436994_a_at 1438204_at | 18453 | -1.146 | -0.4071 | Yes | ||
| 126 | CBX4 | 1419583_at 1440479_at | 18531 | -1.182 | -0.4060 | Yes | ||
| 127 | EPN1 | 1427039_at | 18735 | -1.281 | -0.4103 | Yes | ||
| 128 | ZNF385 | 1418865_at | 18797 | -1.323 | -0.4079 | Yes | ||
| 129 | SLC12A7 | 1418257_at 1449011_at | 18915 | -1.381 | -0.4079 | Yes | ||
| 130 | PCDHGC3 | 1437444_at 1446332_at 1458721_at | 18935 | -1.394 | -0.4033 | Yes | ||
| 131 | SH3BGRL | 1421871_at 1428107_at 1436997_x_at 1459571_at | 18950 | -1.409 | -0.3985 | Yes | ||
| 132 | BBX | 1422741_a_at 1425835_a_at 1430820_a_at 1458602_at | 19016 | -1.440 | -0.3958 | Yes | ||
| 133 | IFI44 | 1423555_a_at | 19095 | -1.497 | -0.3936 | Yes | ||
| 134 | DUSP10 | 1417163_at 1417164_at | 19106 | -1.503 | -0.3882 | Yes | ||
| 135 | PLAG1 | 1421745_at 1443943_at | 19277 | -1.616 | -0.3897 | Yes | ||
| 136 | DAPP1 | 1421936_at 1421937_at 1447337_at | 19606 | -1.880 | -0.3974 | Yes | ||
| 137 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 19804 | -2.024 | -0.3986 | Yes | ||
| 138 | PRKD2 | 1434333_a_at 1434334_at 1436589_x_at 1437509_x_at 1456075_at | 19937 | -2.112 | -0.3964 | Yes | ||
| 139 | HPCAL1 | 1448812_at | 19989 | -2.168 | -0.3902 | Yes | ||
| 140 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 20041 | -2.216 | -0.3839 | Yes | ||
| 141 | CXCR4 | 1448710_at | 20095 | -2.256 | -0.3776 | Yes | ||
| 142 | GRIPAP1 | 1417315_at 1447072_at | 20306 | -2.478 | -0.3775 | Yes | ||
| 143 | CCDC6 | 1428311_at 1459159_a_at | 20344 | -2.525 | -0.3694 | Yes | ||
| 144 | MAP3K11 | 1450669_at | 20460 | -2.667 | -0.3642 | Yes | ||
| 145 | STAT6 | 1421708_a_at 1426353_at | 20465 | -2.672 | -0.3540 | Yes | ||
| 146 | JMJD2A | 1435340_at 1446699_at | 20602 | -2.862 | -0.3491 | Yes | ||
| 147 | FYCO1 | 1427177_at 1453424_at | 20608 | -2.875 | -0.3381 | Yes | ||
| 148 | OSBP | 1460350_at | 20610 | -2.881 | -0.3269 | Yes | ||
| 149 | TCF7L2 | 1425229_a_at 1426639_a_at 1429427_s_at 1429428_at 1441756_at 1446452_at 1447071_at | 20624 | -2.910 | -0.3161 | Yes | ||
| 150 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 20789 | -3.168 | -0.3113 | Yes | ||
| 151 | SERHL | 1421938_at 1434145_s_at 1441234_at 1450419_at | 20908 | -3.399 | -0.3034 | Yes | ||
| 152 | ZBP1 | 1419604_at 1429947_a_at | 20923 | -3.433 | -0.2907 | Yes | ||
| 153 | PML | 1448757_at 1456103_at 1459137_at | 20925 | -3.435 | -0.2773 | Yes | ||
| 154 | RBPSUH | 1418114_at 1438848_at 1446436_at 1448957_at 1454896_at | 20930 | -3.443 | -0.2640 | Yes | ||
| 155 | USP18 | 1418191_at 1444449_at | 21034 | -3.662 | -0.2545 | Yes | ||
| 156 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 21287 | -4.272 | -0.2494 | Yes | ||
| 157 | GPR18 | 1439141_at | 21379 | -4.656 | -0.2354 | Yes | ||
| 158 | FBXO11 | 1434108_at | 21479 | -5.063 | -0.2201 | Yes | ||
| 159 | UBE1L | 1426971_at 1437317_at | 21485 | -5.085 | -0.2005 | Yes | ||
| 160 | CRY1 | 1433733_a_at | 21580 | -5.660 | -0.1827 | Yes | ||
| 161 | ISGF3G | 1421322_a_at | 21588 | -5.690 | -0.1608 | Yes | ||
| 162 | SSBP2 | 1429951_at 1438423_at 1449815_a_at 1451542_at | 21731 | -6.802 | -0.1408 | Yes | ||
| 163 | DGKA | 1418578_at | 21744 | -6.953 | -0.1142 | Yes | ||
| 164 | LSP1 | 1417756_a_at | 21836 | -7.735 | -0.0882 | Yes | ||
| 165 | PPP2R5C | 1425542_a_at 1425725_s_at 1425726_x_at 1427003_at 1434205_at 1434206_s_at 1444828_at | 21843 | -7.790 | -0.0580 | Yes | ||
| 166 | CDKN2C | 1416868_at 1439164_at | 21857 | -7.825 | -0.0281 | Yes | ||
| 167 | EGR2 | 1427682_a_at 1427683_at | 21904 | -8.113 | 0.0015 | Yes |