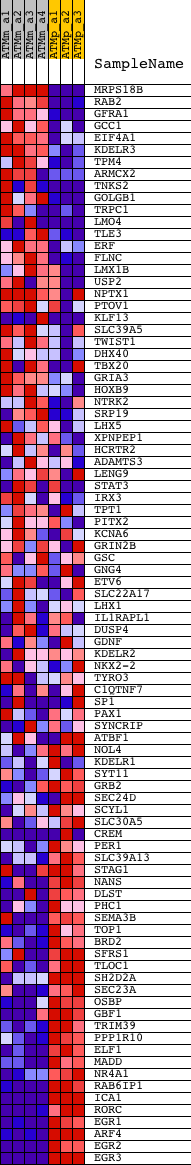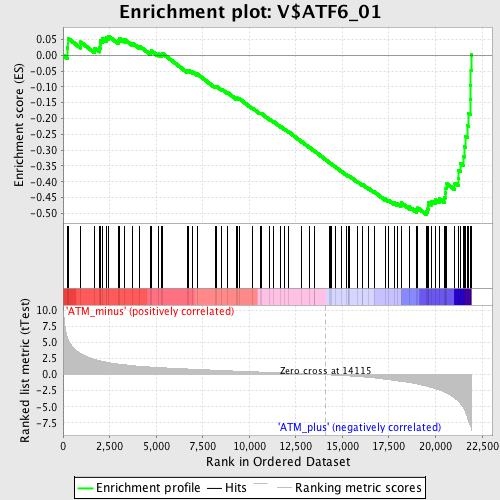
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$ATF6_01 |
| Enrichment Score (ES) | -0.50335586 |
| Normalized Enrichment Score (NES) | -1.8110741 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.026996475 |
| FWER p-Value | 0.055 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MRPS18B | 1451164_a_at | 229 | 5.700 | 0.0231 | No | ||
| 2 | RAB2 | 1418621_at 1418622_at 1418623_at 1419945_s_at 1419946_s_at 1436477_x_at 1449676_at | 262 | 5.426 | 0.0536 | No | ||
| 3 | GFRA1 | 1421973_at 1439015_at 1450440_at | 937 | 3.279 | 0.0420 | No | ||
| 4 | GCC1 | 1424799_a_at 1429033_at | 1681 | 2.402 | 0.0221 | No | ||
| 5 | EIF4A1 | 1427058_at 1430980_a_at 1434985_a_at | 1943 | 2.186 | 0.0231 | No | ||
| 6 | KDELR3 | 1418538_at | 1992 | 2.154 | 0.0335 | No | ||
| 7 | TPM4 | 1433883_at | 1999 | 2.148 | 0.0459 | No | ||
| 8 | ARMCX2 | 1415780_a_at 1437849_x_at 1456739_x_at | 2123 | 2.060 | 0.0524 | No | ||
| 9 | TNKS2 | 1428388_at 1437793_at 1447522_s_at 1452772_at 1457468_at | 2316 | 1.941 | 0.0550 | No | ||
| 10 | GOLGB1 | 1433135_at 1435032_at 1460535_at | 2440 | 1.869 | 0.0604 | No | ||
| 11 | TRPC1 | 1421095_a_at 1421096_at | 2954 | 1.662 | 0.0467 | No | ||
| 12 | LMO4 | 1420981_a_at 1439651_at | 3025 | 1.633 | 0.0531 | No | ||
| 13 | TLE3 | 1419654_at 1419655_at 1449554_at 1458512_at | 3278 | 1.548 | 0.0507 | No | ||
| 14 | ERF | 1422114_at 1435561_at | 3738 | 1.396 | 0.0379 | No | ||
| 15 | FLNC | 1424063_at 1447812_x_at 1449073_at | 4115 | 1.305 | 0.0284 | No | ||
| 16 | LMX1B | 1421769_at | 4709 | 1.190 | 0.0082 | No | ||
| 17 | USP2 | 1417168_a_at 1417169_at | 4736 | 1.184 | 0.0140 | No | ||
| 18 | NPTX1 | 1422130_at 1434877_at | 5098 | 1.116 | 0.0041 | No | ||
| 19 | PTOV1 | 1416945_at 1438058_s_at 1458357_x_at | 5300 | 1.083 | 0.0012 | No | ||
| 20 | KLF13 | 1432543_a_at 1456030_at | 5344 | 1.073 | 0.0056 | No | ||
| 21 | SLC39A5 | 1429523_a_at | 6656 | 0.891 | -0.0492 | No | ||
| 22 | TWIST1 | 1418733_at | 6725 | 0.881 | -0.0471 | No | ||
| 23 | DHX40 | 1428267_at 1457839_at | 6958 | 0.845 | -0.0527 | No | ||
| 24 | TBX20 | 1422219_a_at 1425158_at 1453351_at | 7202 | 0.812 | -0.0591 | No | ||
| 25 | GRIA3 | 1420563_at 1434728_at | 8160 | 0.687 | -0.0988 | No | ||
| 26 | HOXB9 | 1452317_at | 8253 | 0.676 | -0.0991 | No | ||
| 27 | NTRK2 | 1420837_at 1420838_at 1435196_at 1435305_at 1437560_at 1446712_at | 8510 | 0.644 | -0.1070 | No | ||
| 28 | SRP19 | 1450891_at | 8846 | 0.604 | -0.1188 | No | ||
| 29 | LHX5 | 1420348_at | 9333 | 0.547 | -0.1378 | No | ||
| 30 | XPNPEP1 | 1422443_at | 9343 | 0.546 | -0.1350 | No | ||
| 31 | HCRTR2 | 1444223_at | 9470 | 0.531 | -0.1376 | No | ||
| 32 | ADAMTS3 | 1441693_at 1458143_at | 10171 | 0.451 | -0.1670 | No | ||
| 33 | LENG9 | 1440190_at 1440191_s_at 1455497_at | 10579 | 0.411 | -0.1832 | No | ||
| 34 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 10629 | 0.405 | -0.1831 | No | ||
| 35 | IRX3 | 1418517_at | 11105 | 0.350 | -0.2028 | No | ||
| 36 | TPT1 | 1416642_a_at 1416643_at | 11317 | 0.326 | -0.2105 | No | ||
| 37 | PITX2 | 1424797_a_at 1450482_a_at | 11687 | 0.286 | -0.2257 | No | ||
| 38 | KCNA6 | 1421568_at 1441049_at 1456954_at | 11885 | 0.263 | -0.2332 | No | ||
| 39 | GRIN2B | 1422223_at 1431700_at | 12124 | 0.239 | -0.2427 | No | ||
| 40 | GSC | 1421412_at | 12822 | 0.163 | -0.2736 | No | ||
| 41 | GNG4 | 1417943_at 1417944_at 1447669_s_at | 13231 | 0.115 | -0.2916 | No | ||
| 42 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 13474 | 0.085 | -0.3022 | No | ||
| 43 | SLC22A17 | 1448209_a_at | 13504 | 0.081 | -0.3031 | No | ||
| 44 | LHX1 | 1421951_at 1450428_at | 14310 | -0.030 | -0.3397 | No | ||
| 45 | IL1RAPL1 | 1445821_at 1458964_at | 14354 | -0.036 | -0.3415 | No | ||
| 46 | DUSP4 | 1428834_at | 14418 | -0.045 | -0.3441 | No | ||
| 47 | GDNF | 1419080_at | 14652 | -0.083 | -0.3543 | No | ||
| 48 | KDELR2 | 1417204_at 1417205_at | 14962 | -0.132 | -0.3677 | No | ||
| 49 | NKX2-2 | 1421112_at | 15245 | -0.177 | -0.3795 | No | ||
| 50 | TYRO3 | 1425248_a_at 1425249_a_at | 15300 | -0.185 | -0.3809 | No | ||
| 51 | C1QTNF7 | 1429030_at 1460163_at | 15315 | -0.188 | -0.3804 | No | ||
| 52 | SP1 | 1418180_at 1448994_at 1454852_at | 15376 | -0.199 | -0.3820 | No | ||
| 53 | PAX1 | 1449359_at | 15819 | -0.285 | -0.4006 | No | ||
| 54 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 16053 | -0.337 | -0.4092 | No | ||
| 55 | ATBF1 | 1420649_at 1420650_at 1429725_at 1445941_at 1446094_at 1449947_s_at 1453267_at 1460006_at | 16065 | -0.340 | -0.4077 | No | ||
| 56 | NOL4 | 1430189_at 1441447_at 1456387_at | 16406 | -0.421 | -0.4208 | No | ||
| 57 | KDELR1 | 1460211_a_at | 16701 | -0.498 | -0.4313 | No | ||
| 58 | SYT11 | 1429314_at 1429315_at 1429729_at 1436176_at 1449264_at 1455176_a_at | 17287 | -0.690 | -0.4541 | No | ||
| 59 | GRB2 | 1418508_a_at 1449111_a_at | 17468 | -0.764 | -0.4578 | No | ||
| 60 | SEC24D | 1426972_at 1458821_at | 17776 | -0.893 | -0.4666 | No | ||
| 61 | SCYL1 | 1430354_x_at 1436804_s_at 1451051_a_at | 17947 | -0.963 | -0.4687 | No | ||
| 62 | SLC30A5 | 1422497_at | 18149 | -1.024 | -0.4719 | No | ||
| 63 | CREM | 1418322_at 1430598_at 1430847_a_at 1449037_at | 18158 | -1.026 | -0.4662 | No | ||
| 64 | PER1 | 1449851_at | 18601 | -1.214 | -0.4793 | No | ||
| 65 | SLC39A13 | 1423926_at | 18970 | -1.419 | -0.4878 | No | ||
| 66 | STAG1 | 1421939_a_at 1421940_at 1431921_a_at 1434189_at 1435731_x_at 1445145_at 1445652_at 1450420_at | 19049 | -1.462 | -0.4828 | No | ||
| 67 | NANS | 1417773_at 1417774_at | 19500 | -1.772 | -0.4929 | Yes | ||
| 68 | DLST | 1423710_at 1430161_at 1431011_at 1437775_at | 19585 | -1.857 | -0.4858 | Yes | ||
| 69 | PHC1 | 1423212_at | 19605 | -1.878 | -0.4756 | Yes | ||
| 70 | SEMA3B | 1416644_a_at 1431795_a_at 1448415_a_at | 19610 | -1.885 | -0.4647 | Yes | ||
| 71 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 19804 | -2.024 | -0.4617 | Yes | ||
| 72 | BRD2 | 1423502_at 1437210_a_at | 19987 | -2.166 | -0.4572 | Yes | ||
| 73 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 20198 | -2.362 | -0.4530 | Yes | ||
| 74 | TLOC1 | 1432635_a_at 1433704_s_at 1439233_at 1444811_at 1454697_at 1455698_at | 20491 | -2.716 | -0.4503 | Yes | ||
| 75 | SH2D2A | 1449105_at | 20515 | -2.759 | -0.4351 | Yes | ||
| 76 | SEC23A | 1423347_at | 20559 | -2.820 | -0.4205 | Yes | ||
| 77 | OSBP | 1460350_at | 20610 | -2.881 | -0.4058 | Yes | ||
| 78 | GBF1 | 1415709_s_at 1415757_at 1432677_at 1438207_at | 21042 | -3.685 | -0.4039 | Yes | ||
| 79 | TRIM39 | 1418851_at | 21234 | -4.127 | -0.3883 | Yes | ||
| 80 | PPP1R10 | 1426726_at 1426727_s_at 1430560_at | 21251 | -4.166 | -0.3645 | Yes | ||
| 81 | ELF1 | 1417540_at 1439319_at | 21332 | -4.495 | -0.3417 | Yes | ||
| 82 | MADD | 1425735_at 1455502_at | 21500 | -5.159 | -0.3190 | Yes | ||
| 83 | NR4A1 | 1416505_at | 21561 | -5.542 | -0.2891 | Yes | ||
| 84 | RAB6IP1 | 1424015_at | 21629 | -5.995 | -0.2569 | Yes | ||
| 85 | ICA1 | 1417901_a_at 1431644_a_at | 21702 | -6.525 | -0.2218 | Yes | ||
| 86 | RORC | 1425792_a_at 1425793_a_at | 21785 | -7.272 | -0.1827 | Yes | ||
| 87 | EGR1 | 1417065_at | 21860 | -7.832 | -0.1400 | Yes | ||
| 88 | ARF4 | 1423052_at 1423053_at 1434074_x_at 1439367_x_at 1442905_at 1457348_at | 21892 | -7.985 | -0.0944 | Yes | ||
| 89 | EGR2 | 1427682_a_at 1427683_at | 21904 | -8.113 | -0.0472 | Yes | ||
| 90 | EGR3 | 1421486_at 1436329_at | 21913 | -8.258 | 0.0011 | Yes |