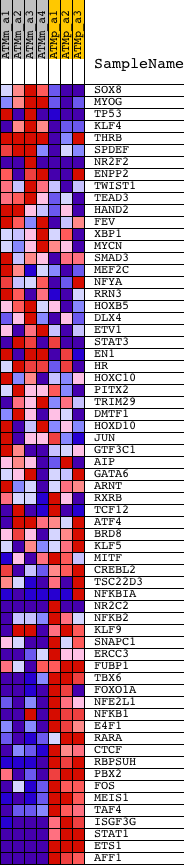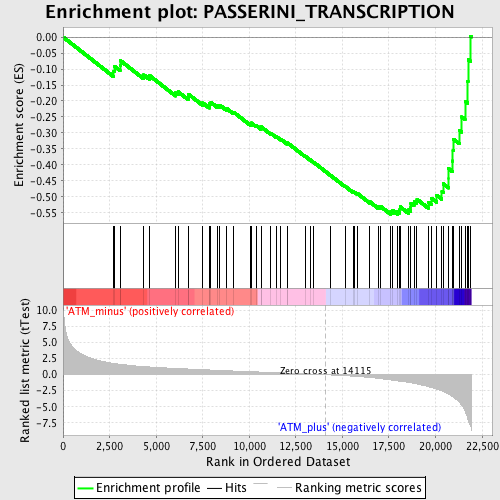
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | PASSERINI_TRANSCRIPTION |
| Enrichment Score (ES) | -0.5549399 |
| Normalized Enrichment Score (NES) | -1.9102993 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.034921084 |
| FWER p-Value | 0.243 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SOX8 | 1435438_at | 2723 | 1.741 | -0.1071 | No | ||
| 2 | MYOG | 1419391_at | 2761 | 1.730 | -0.0915 | No | ||
| 3 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 3061 | 1.619 | -0.0890 | No | ||
| 4 | KLF4 | 1417394_at 1417395_at | 3080 | 1.612 | -0.0738 | No | ||
| 5 | THRB | 1422202_at 1443100_at | 4335 | 1.261 | -0.1185 | No | ||
| 6 | SPDEF | 1450220_a_at | 4643 | 1.203 | -0.1205 | No | ||
| 7 | NR2F2 | 1416158_at 1416159_at 1416160_at 1431237_at 1444229_at | 6042 | 0.981 | -0.1746 | No | ||
| 8 | ENPP2 | 1415894_at 1443224_at 1448136_at | 6180 | 0.960 | -0.1713 | No | ||
| 9 | TWIST1 | 1418733_at | 6725 | 0.881 | -0.1874 | No | ||
| 10 | TEAD3 | 1420593_a_at | 6733 | 0.880 | -0.1789 | No | ||
| 11 | HAND2 | 1422221_at 1436041_at 1443854_at 1446812_at | 7496 | 0.776 | -0.2060 | No | ||
| 12 | FEV | 1425886_at | 7847 | 0.730 | -0.2147 | No | ||
| 13 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 7859 | 0.728 | -0.2080 | No | ||
| 14 | MYCN | 1417155_at 1425922_a_at 1425923_at | 7938 | 0.720 | -0.2044 | No | ||
| 15 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 8283 | 0.672 | -0.2134 | No | ||
| 16 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 8419 | 0.655 | -0.2130 | No | ||
| 17 | NFYA | 1422082_a_at 1427808_at 1452560_a_at | 8795 | 0.611 | -0.2241 | No | ||
| 18 | RRN3 | 1415725_at | 9156 | 0.567 | -0.2348 | No | ||
| 19 | HOXB5 | 1418415_at | 10060 | 0.462 | -0.2715 | No | ||
| 20 | DLX4 | 1420595_at 1427641_at 1443591_at | 10099 | 0.458 | -0.2687 | No | ||
| 21 | ETV1 | 1422607_at 1445314_at 1450684_at | 10374 | 0.431 | -0.2769 | No | ||
| 22 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 10629 | 0.405 | -0.2845 | No | ||
| 23 | EN1 | 1418618_at | 10639 | 0.404 | -0.2809 | No | ||
| 24 | HR | 1422110_at 1435950_at | 11152 | 0.346 | -0.3008 | No | ||
| 25 | HOXC10 | 1439798_at | 11465 | 0.307 | -0.3120 | No | ||
| 26 | PITX2 | 1424797_a_at 1450482_a_at | 11687 | 0.286 | -0.3193 | No | ||
| 27 | TRIM29 | 1424162_at | 12045 | 0.247 | -0.3331 | No | ||
| 28 | DMTF1 | 1422636_at 1440349_at 1456697_x_at | 12052 | 0.246 | -0.3310 | No | ||
| 29 | HOXD10 | 1418606_at | 12998 | 0.142 | -0.3727 | No | ||
| 30 | JUN | 1417409_at 1448694_at | 13265 | 0.110 | -0.3838 | No | ||
| 31 | GTF3C1 | 1434066_at 1438425_at 1456024_at 1459673_at | 13459 | 0.086 | -0.3918 | No | ||
| 32 | AIP | 1423128_at | 14378 | -0.040 | -0.4333 | No | ||
| 33 | GATA6 | 1425463_at 1425464_at 1443081_at | 15142 | -0.160 | -0.4666 | No | ||
| 34 | ARNT | 1419996_s_at 1421721_a_at 1425064_at 1437042_at 1449696_at | 15570 | -0.233 | -0.4838 | No | ||
| 35 | RXRB | 1416990_at | 15644 | -0.248 | -0.4847 | No | ||
| 36 | TCF12 | 1421908_a_at 1427670_a_at 1438762_at 1439209_at 1439619_at 1443012_at 1444620_at 1445093_at 1445133_at 1457524_at 1458337_at | 15793 | -0.280 | -0.4887 | No | ||
| 37 | ATF4 | 1438992_x_at 1439258_at 1448135_at | 16468 | -0.438 | -0.5151 | No | ||
| 38 | BRD8 | 1427192_a_at 1427193_at 1429526_at 1451137_a_at 1452350_at 1456351_at | 16942 | -0.574 | -0.5310 | No | ||
| 39 | KLF5 | 1430255_at 1451021_a_at 1451739_at | 17065 | -0.615 | -0.5304 | No | ||
| 40 | MITF | 1422025_at 1455214_at | 17602 | -0.821 | -0.5467 | Yes | ||
| 41 | CREBL2 | 1435085_at 1442738_at | 17700 | -0.858 | -0.5426 | Yes | ||
| 42 | TSC22D3 | 1420772_a_at 1425281_a_at 1425557_x_at 1444365_at | 17954 | -0.965 | -0.5445 | Yes | ||
| 43 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 18087 | -1.008 | -0.5405 | Yes | ||
| 44 | NR2C2 | 1425014_at 1439696_at 1445613_at 1446955_at 1451569_at 1454167_at 1454851_at | 18092 | -1.009 | -0.5306 | Yes | ||
| 45 | NFKB2 | 1425902_a_at 1429128_x_at | 18571 | -1.202 | -0.5405 | Yes | ||
| 46 | KLF9 | 1422264_s_at 1428288_at 1428289_at 1436763_a_at 1436952_at 1452723_at 1456341_a_at | 18672 | -1.248 | -0.5326 | Yes | ||
| 47 | SNAPC1 | 1429335_at | 18678 | -1.249 | -0.5203 | Yes | ||
| 48 | ERCC3 | 1448497_at | 18848 | -1.346 | -0.5146 | Yes | ||
| 49 | FUBP1 | 1433482_a_at 1433640_at 1437543_at 1437544_at | 19003 | -1.434 | -0.5073 | Yes | ||
| 50 | TBX6 | 1449868_at | 19608 | -1.881 | -0.5162 | Yes | ||
| 51 | FOXO1A | 1416981_at 1416982_at 1416983_s_at 1459170_at | 19785 | -2.010 | -0.5042 | Yes | ||
| 52 | NFE2L1 | 1416331_a_at | 20066 | -2.236 | -0.4946 | Yes | ||
| 53 | NFKB1 | 1427705_a_at 1442949_at | 20346 | -2.528 | -0.4822 | Yes | ||
| 54 | E4F1 | 1450464_at | 20416 | -2.622 | -0.4591 | Yes | ||
| 55 | RARA | 1450180_a_at | 20702 | -3.029 | -0.4419 | Yes | ||
| 56 | CTCF | 1418330_at 1449042_at | 20704 | -3.033 | -0.4117 | Yes | ||
| 57 | RBPSUH | 1418114_at 1438848_at 1446436_at 1448957_at 1454896_at | 20930 | -3.443 | -0.3876 | Yes | ||
| 58 | PBX2 | 1418893_at 1418894_s_at 1449261_at | 20938 | -3.463 | -0.3533 | Yes | ||
| 59 | FOS | 1423100_at | 20955 | -3.496 | -0.3191 | Yes | ||
| 60 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 21287 | -4.272 | -0.2916 | Yes | ||
| 61 | TAF4 | 1427460_at 1452438_s_at | 21414 | -4.759 | -0.2499 | Yes | ||
| 62 | ISGF3G | 1421322_a_at | 21588 | -5.690 | -0.2010 | Yes | ||
| 63 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 21734 | -6.804 | -0.1397 | Yes | ||
| 64 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 21749 | -7.027 | -0.0701 | Yes | ||
| 65 | AFF1 | 1418135_at 1425640_at 1425641_at 1441594_at 1444937_at | 21873 | -7.875 | 0.0029 | Yes |