Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
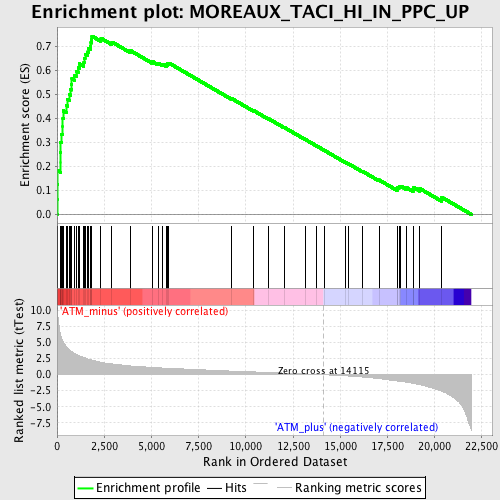
| GeneSet | MOREAUX_TACI_HI_IN_PPC_UP |
| Enrichment Score (ES) | 0.74357635 |
| Normalized Enrichment Score (NES) | 2.4668152 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

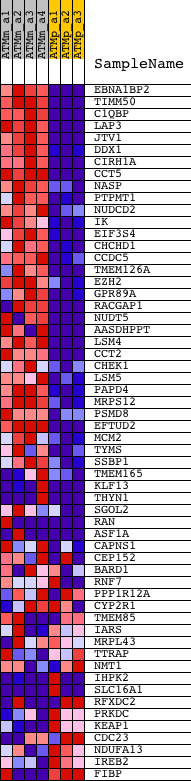
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | EBNA1BP2 | 1428315_at 1437512_x_at 1439898_at 1457668_x_at | 7 | 9.579 | 0.0627 | Yes | ||
| 2 | TIMM50 | 1441819_x_at 1455456_a_at | 14 | 9.397 | 0.1242 | Yes | ||
| 3 | C1QBP | 1448274_at 1455821_x_at | 41 | 8.945 | 0.1819 | Yes | ||
| 4 | LAP3 | 1450860_at | 168 | 6.388 | 0.2182 | Yes | ||
| 5 | JTV1 | 1424151_at 1451262_a_at | 169 | 6.382 | 0.2602 | Yes | ||
| 6 | DDX1 | 1415915_at 1440816_x_at | 179 | 6.206 | 0.3006 | Yes | ||
| 7 | CIRH1A | 1423884_at | 234 | 5.643 | 0.3352 | Yes | ||
| 8 | CCT5 | 1417258_at | 303 | 5.162 | 0.3661 | Yes | ||
| 9 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 307 | 5.145 | 0.3998 | Yes | ||
| 10 | PTPMT1 | 1426581_at | 329 | 5.056 | 0.4321 | Yes | ||
| 11 | NUDCD2 | 1418566_s_at 1449140_at | 482 | 4.414 | 0.4542 | Yes | ||
| 12 | IK | 1423791_at 1435988_x_at 1439271_x_at 1451113_a_at | 543 | 4.203 | 0.4791 | Yes | ||
| 13 | EIF3S4 | 1417718_at | 668 | 3.863 | 0.4989 | Yes | ||
| 14 | CHCHD1 | 1416938_at | 721 | 3.725 | 0.5210 | Yes | ||
| 15 | CCDC5 | 1424955_at | 770 | 3.617 | 0.5426 | Yes | ||
| 16 | TMEM126A | 1451000_at | 784 | 3.584 | 0.5656 | Yes | ||
| 17 | EZH2 | 1416544_at 1444263_at | 932 | 3.290 | 0.5805 | Yes | ||
| 18 | GPR89A | 1424696_at | 1034 | 3.120 | 0.5964 | Yes | ||
| 19 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 1142 | 2.963 | 0.6110 | Yes | ||
| 20 | NUDT5 | 1430341_at 1448651_at | 1187 | 2.916 | 0.6282 | Yes | ||
| 21 | AASDHPPT | 1428756_at 1428757_at 1430515_s_at | 1418 | 2.679 | 0.6353 | Yes | ||
| 22 | LSM4 | 1448622_at | 1431 | 2.661 | 0.6523 | Yes | ||
| 23 | CCT2 | 1416037_a_at 1433534_a_at 1433535_x_at | 1480 | 2.613 | 0.6673 | Yes | ||
| 24 | CHEK1 | 1420031_at 1420032_at 1439208_at 1449708_s_at 1450677_at | 1615 | 2.468 | 0.6774 | Yes | ||
| 25 | LSM5 | 1418656_at | 1669 | 2.412 | 0.6908 | Yes | ||
| 26 | PAPD4 | 1419183_at 1433568_at | 1765 | 2.333 | 0.7018 | Yes | ||
| 27 | MRPS12 | 1448799_s_at | 1791 | 2.316 | 0.7159 | Yes | ||
| 28 | PSMD8 | 1423296_at | 1817 | 2.287 | 0.7298 | Yes | ||
| 29 | EFTUD2 | 1416557_a_at 1438835_a_at 1438836_at 1456107_x_at | 1843 | 2.263 | 0.7436 | Yes | ||
| 30 | MCM2 | 1434079_s_at 1448777_at | 2322 | 1.939 | 0.7345 | No | ||
| 31 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 2905 | 1.680 | 0.7189 | No | ||
| 32 | SSBP1 | 1427965_at 1430294_at 1444053_at 1452616_s_at 1452617_at | 3881 | 1.361 | 0.6833 | No | ||
| 33 | TMEM165 | 1415741_at | 5057 | 1.122 | 0.6370 | No | ||
| 34 | KLF13 | 1432543_a_at 1456030_at | 5344 | 1.073 | 0.6310 | No | ||
| 35 | THYN1 | 1438480_a_at 1438769_a_at 1460007_at | 5602 | 1.037 | 0.6261 | No | ||
| 36 | SGOL2 | 1437370_at | 5786 | 1.011 | 0.6244 | No | ||
| 37 | RAN | 1443131_at 1446819_at 1447478_at 1460551_at | 5842 | 1.004 | 0.6284 | No | ||
| 38 | ASF1A | 1423511_at 1459882_at | 5921 | 0.997 | 0.6314 | No | ||
| 39 | CAPNS1 | 1426400_a_at | 9228 | 0.559 | 0.4840 | No | ||
| 40 | CEP152 | 1427496_at | 10406 | 0.427 | 0.4330 | No | ||
| 41 | BARD1 | 1420594_at | 11209 | 0.338 | 0.3986 | No | ||
| 42 | RNF7 | 1426414_a_at 1456600_a_at | 12021 | 0.249 | 0.3632 | No | ||
| 43 | PPP1R12A | 1429487_at 1437734_at 1437735_at 1444762_at 1444834_at 1453163_at | 13142 | 0.124 | 0.3128 | No | ||
| 44 | CYP2R1 | 1444138_at | 13760 | 0.045 | 0.2849 | No | ||
| 45 | TMEM85 | 1418124_at 1438369_x_at | 14135 | -0.002 | 0.2678 | No | ||
| 46 | IARS | 1426705_s_at 1452154_at | 15256 | -0.178 | 0.2178 | No | ||
| 47 | MRPL43 | 1415681_at | 15423 | -0.206 | 0.2116 | No | ||
| 48 | TTRAP | 1448706_at | 16149 | -0.358 | 0.1808 | No | ||
| 49 | NMT1 | 1415683_at 1418082_at 1454941_at | 17050 | -0.610 | 0.1437 | No | ||
| 50 | IHPK2 | 1428373_at 1435319_at | 18009 | -0.985 | 0.1064 | No | ||
| 51 | SLC16A1 | 1415802_at 1446274_at | 18010 | -0.986 | 0.1128 | No | ||
| 52 | RFXDC2 | 1434521_at 1440068_at 1457847_at 1460567_at | 18109 | -1.013 | 0.1150 | No | ||
| 53 | PRKDC | 1451576_at | 18211 | -1.043 | 0.1173 | No | ||
| 54 | KEAP1 | 1450746_at 1450747_at | 18520 | -1.178 | 0.1109 | No | ||
| 55 | CDC23 | 1433430_s_at 1443087_at 1460549_a_at | 18856 | -1.350 | 0.1045 | No | ||
| 56 | NDUFA13 | 1430713_s_at | 18872 | -1.359 | 0.1128 | No | ||
| 57 | IREB2 | 1433527_at 1458618_at | 19213 | -1.576 | 0.1076 | No | ||
| 58 | FIBP | 1422516_a_at 1430171_at 1438923_at 1438924_x_at | 20368 | -2.556 | 0.0717 | No |