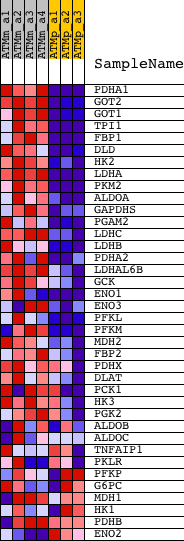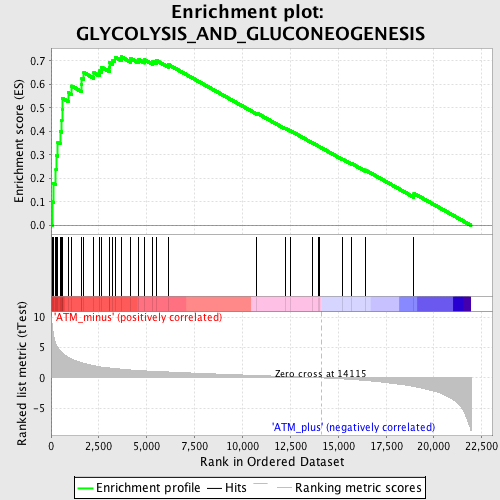
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | 0.7170504 |
| Normalized Enrichment Score (NES) | 2.222796 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PDHA1 | 1418560_at 1446954_at 1449137_at | 52 | 8.811 | 0.1008 | Yes | ||
| 2 | GOT2 | 1417715_a_at 1417716_at | 123 | 6.994 | 0.1795 | Yes | ||
| 3 | GOT1 | 1450970_at | 252 | 5.481 | 0.2378 | Yes | ||
| 4 | TPI1 | 1415918_a_at 1435659_a_at 1452927_x_at | 290 | 5.245 | 0.2975 | Yes | ||
| 5 | FBP1 | 1448470_at | 353 | 4.968 | 0.3529 | Yes | ||
| 6 | DLD | 1423159_at | 470 | 4.457 | 0.3998 | Yes | ||
| 7 | HK2 | 1422612_at | 550 | 4.184 | 0.4452 | Yes | ||
| 8 | LDHA | 1419737_a_at | 570 | 4.137 | 0.4927 | Yes | ||
| 9 | PKM2 | 1417308_at | 576 | 4.123 | 0.5408 | Yes | ||
| 10 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 913 | 3.316 | 0.5642 | Yes | ||
| 11 | GAPDHS | 1449974_at | 1070 | 3.071 | 0.5931 | Yes | ||
| 12 | PGAM2 | 1418373_at | 1596 | 2.487 | 0.5982 | Yes | ||
| 13 | LDHC | 1415846_a_at 1415847_at | 1606 | 2.478 | 0.6268 | Yes | ||
| 14 | LDHB | 1416183_a_at 1434499_a_at 1442618_at 1448237_x_at 1455235_x_at | 1713 | 2.375 | 0.6498 | Yes | ||
| 15 | PDHA2 | 1450962_at | 2234 | 1.990 | 0.6494 | Yes | ||
| 16 | LDHAL6B | 1434247_at | 2532 | 1.820 | 0.6571 | Yes | ||
| 17 | GCK | 1419146_a_at 1425303_at | 2643 | 1.772 | 0.6728 | Yes | ||
| 18 | ENO1 | 1419023_x_at | 3045 | 1.627 | 0.6736 | Yes | ||
| 19 | ENO3 | 1417951_at 1449631_at | 3066 | 1.617 | 0.6916 | Yes | ||
| 20 | PFKL | 1439148_a_at 1450269_a_at 1453541_at | 3226 | 1.560 | 0.7026 | Yes | ||
| 21 | PFKM | 1416780_at | 3337 | 1.529 | 0.7155 | Yes | ||
| 22 | MDH2 | 1416478_a_at 1433984_a_at | 3667 | 1.418 | 0.7171 | Yes | ||
| 23 | FBP2 | 1449088_at | 4157 | 1.296 | 0.7099 | No | ||
| 24 | PDHX | 1456090_at 1459434_at | 4561 | 1.220 | 0.7058 | No | ||
| 25 | DLAT | 1426264_at 1426265_x_at 1452005_at | 4888 | 1.155 | 0.7044 | No | ||
| 26 | PCK1 | 1423439_at 1439617_s_at 1455209_at | 5293 | 1.084 | 0.6987 | No | ||
| 27 | HK3 | 1435490_at 1442798_x_at | 5505 | 1.048 | 0.7013 | No | ||
| 28 | PGK2 | 1416784_at | 6127 | 0.968 | 0.6843 | No | ||
| 29 | ALDOB | 1451194_at | 10747 | 0.390 | 0.4779 | No | ||
| 30 | ALDOC | 1424714_at 1451461_a_at | 12229 | 0.228 | 0.4130 | No | ||
| 31 | TNFAIP1 | 1417865_at 1417866_at 1448863_a_at | 12490 | 0.200 | 0.4034 | No | ||
| 32 | PKLR | 1421258_a_at 1421259_at 1438711_at | 13676 | 0.057 | 0.3500 | No | ||
| 33 | PFKP | 1416069_at 1430634_a_at 1437759_at | 13958 | 0.020 | 0.3374 | No | ||
| 34 | G6PC | 1417880_at | 14035 | 0.011 | 0.3340 | No | ||
| 35 | MDH1 | 1434319_at 1436834_x_at 1438338_at 1448172_at 1454925_x_at | 15234 | -0.175 | 0.2814 | No | ||
| 36 | HK1 | 1420901_a_at 1437974_a_at | 15706 | -0.261 | 0.2629 | No | ||
| 37 | PDHB | 1416090_at 1448214_at | 16411 | -0.423 | 0.2357 | No | ||
| 38 | ENO2 | 1418829_a_at | 18955 | -1.410 | 0.1361 | No |