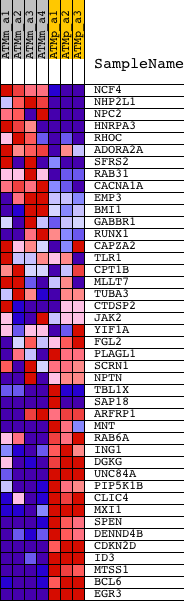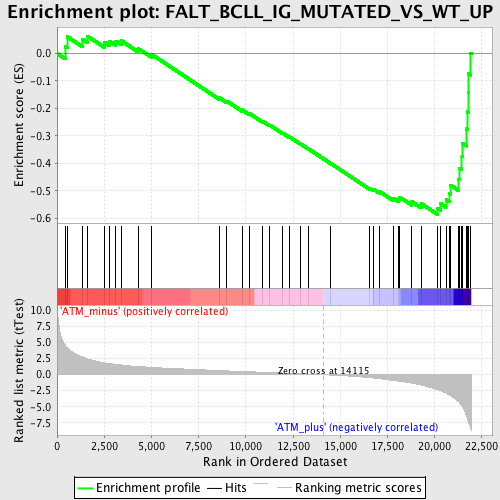
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | FALT_BCLL_IG_MUTATED_VS_WT_UP |
| Enrichment Score (ES) | -0.5862576 |
| Normalized Enrichment Score (NES) | -1.8783801 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.038763184 |
| FWER p-Value | 0.389 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NCF4 | 1418465_at | 440 | 4.586 | 0.0245 | No | ||
| 2 | NHP2L1 | 1416973_at | 534 | 4.229 | 0.0615 | No | ||
| 3 | NPC2 | 1416901_at 1443266_at 1448513_a_at 1455768_at | 1347 | 2.746 | 0.0511 | No | ||
| 4 | HNRPA3 | 1452712_at 1452774_at | 1611 | 2.475 | 0.0632 | No | ||
| 5 | RHOC | 1435394_s_at 1448605_at | 2506 | 1.832 | 0.0402 | No | ||
| 6 | ADORA2A | 1427519_at 1460710_at | 2763 | 1.730 | 0.0453 | No | ||
| 7 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 3110 | 1.602 | 0.0451 | No | ||
| 8 | RAB31 | 1416165_at 1431691_a_at | 3412 | 1.504 | 0.0460 | No | ||
| 9 | CACNA1A | 1430408_at 1444455_at 1450510_a_at 1458312_at 1459996_at | 4297 | 1.269 | 0.0180 | No | ||
| 10 | EMP3 | 1417104_at | 5020 | 1.128 | -0.0040 | No | ||
| 11 | BMI1 | 1417493_at 1448733_at | 8595 | 0.634 | -0.1611 | No | ||
| 12 | GABBR1 | 1422051_a_at 1425595_at 1437188_at 1455021_at | 8985 | 0.588 | -0.1732 | No | ||
| 13 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 9797 | 0.494 | -0.2054 | No | ||
| 14 | CAPZA2 | 1423057_at 1423058_at | 10178 | 0.451 | -0.2184 | No | ||
| 15 | TLR1 | 1449049_at | 10894 | 0.373 | -0.2474 | No | ||
| 16 | CPT1B | 1418328_at | 11255 | 0.333 | -0.2606 | No | ||
| 17 | MLLT7 | 1422149_at | 11962 | 0.255 | -0.2904 | No | ||
| 18 | TUBA3 | 1438611_at | 12309 | 0.221 | -0.3040 | No | ||
| 19 | CTDSP2 | 1423660_at 1423661_s_at 1451075_s_at | 12882 | 0.155 | -0.3286 | No | ||
| 20 | JAK2 | 1421065_at 1421066_at | 13318 | 0.103 | -0.3475 | No | ||
| 21 | YIF1A | 1423939_a_at 1423940_at 1456134_x_at | 14460 | -0.052 | -0.3991 | No | ||
| 22 | FGL2 | 1421854_at 1421855_at | 16545 | -0.455 | -0.4899 | No | ||
| 23 | PLAGL1 | 1426208_x_at 1438588_at 1450533_a_at | 16749 | -0.513 | -0.4941 | No | ||
| 24 | SCRN1 | 1428717_at 1428718_at | 17057 | -0.611 | -0.5022 | No | ||
| 25 | NPTN | 1415821_at 1443037_at | 17816 | -0.912 | -0.5280 | No | ||
| 26 | TBL1X | 1434643_at 1434644_at 1441435_at 1446736_at 1455042_at | 18070 | -1.005 | -0.5297 | No | ||
| 27 | SAP18 | 1419443_at 1419444_at 1419445_s_at 1434646_s_at 1449480_at | 18155 | -1.026 | -0.5236 | No | ||
| 28 | ARFRP1 | 1425507_at 1425508_s_at 1434488_at 1451735_at | 18790 | -1.319 | -0.5397 | No | ||
| 29 | MNT | 1418192_at | 19289 | -1.619 | -0.5467 | No | ||
| 30 | RAB6A | 1447776_x_at 1448304_a_at 1448305_at | 20156 | -2.318 | -0.5637 | Yes | ||
| 31 | ING1 | 1416860_s_at 1433267_at 1448496_a_at 1454534_at | 20323 | -2.498 | -0.5470 | Yes | ||
| 32 | DGKG | 1419756_at 1431167_at 1449584_at | 20599 | -2.861 | -0.5317 | Yes | ||
| 33 | UNC84A | 1426666_a_at 1426667_a_at | 20794 | -3.177 | -0.5096 | Yes | ||
| 34 | PIP5K1B | 1418144_a_at 1426009_a_at | 20847 | -3.288 | -0.4800 | Yes | ||
| 35 | CLIC4 | 1423392_at 1423393_at 1438606_a_at | 21270 | -4.217 | -0.4582 | Yes | ||
| 36 | MXI1 | 1425732_a_at 1450376_at | 21325 | -4.477 | -0.4171 | Yes | ||
| 37 | SPEN | 1420397_a_at | 21443 | -4.872 | -0.3750 | Yes | ||
| 38 | DENND4B | 1427014_at | 21470 | -5.026 | -0.3273 | Yes | ||
| 39 | CDKN2D | 1416253_at | 21682 | -6.381 | -0.2748 | Yes | ||
| 40 | ID3 | 1416630_at | 21719 | -6.663 | -0.2116 | Yes | ||
| 41 | MTSS1 | 1424826_s_at 1434036_at 1440847_at 1443729_at 1446284_at 1451496_at | 21776 | -7.245 | -0.1437 | Yes | ||
| 42 | BCL6 | 1421818_at 1450381_a_at | 21780 | -7.247 | -0.0733 | Yes | ||
| 43 | EGR3 | 1421486_at 1436329_at | 21913 | -8.258 | 0.0011 | Yes |