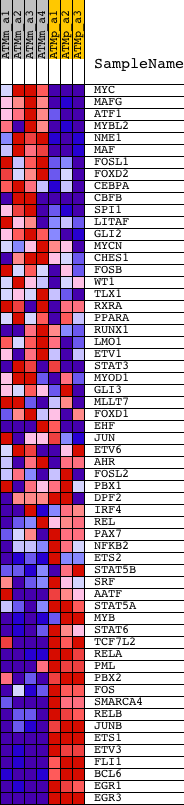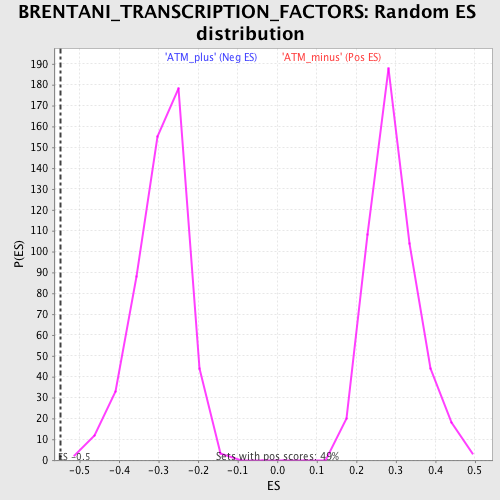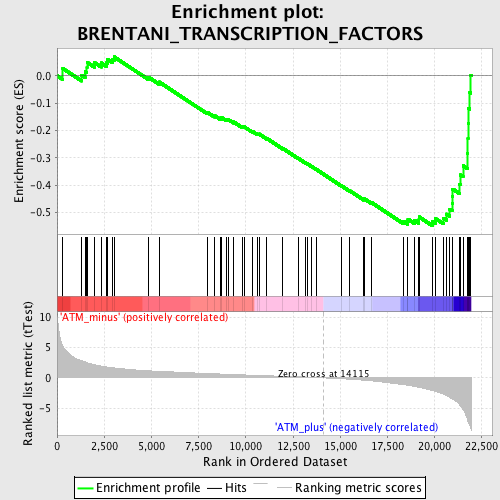
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | BRENTANI_TRANSCRIPTION_FACTORS |
| Enrichment Score (ES) | -0.54844606 |
| Normalized Enrichment Score (NES) | -1.865214 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.04107461 |
| FWER p-Value | 0.444 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MYC | 1424942_a_at | 302 | 5.162 | 0.0265 | No | ||
| 2 | MAFG | 1434054_at 1448916_at | 1301 | 2.793 | 0.0026 | No | ||
| 3 | ATF1 | 1417296_at | 1483 | 2.611 | 0.0147 | No | ||
| 4 | MYBL2 | 1417656_at 1454946_at | 1581 | 2.500 | 0.0298 | No | ||
| 5 | NME1 | 1424110_a_at 1435277_x_at | 1586 | 2.494 | 0.0490 | No | ||
| 6 | MAF | 1435828_at 1437473_at 1444073_at 1447849_s_at 1447945_at 1456060_at | 1970 | 2.170 | 0.0484 | No | ||
| 7 | FOSL1 | 1417487_at 1417488_at | 2333 | 1.933 | 0.0470 | No | ||
| 8 | FOXD2 | 1450289_at | 2635 | 1.775 | 0.0471 | No | ||
| 9 | CEBPA | 1418982_at | 2666 | 1.762 | 0.0594 | No | ||
| 10 | CBFB | 1442617_at 1460716_a_at | 2928 | 1.671 | 0.0605 | No | ||
| 11 | SPI1 | 1418747_at | 3021 | 1.634 | 0.0691 | No | ||
| 12 | LITAF | 1416303_at 1416304_at | 4846 | 1.164 | -0.0052 | No | ||
| 13 | GLI2 | 1446086_s_at 1459211_at | 5408 | 1.063 | -0.0226 | No | ||
| 14 | MYCN | 1417155_at 1425922_a_at 1425923_at | 7938 | 0.720 | -0.1326 | No | ||
| 15 | CHES1 | 1434002_at 1436925_at 1438255_at 1440607_at 1444555_at 1447325_at 1453094_at | 8335 | 0.665 | -0.1455 | No | ||
| 16 | FOSB | 1422134_at | 8652 | 0.626 | -0.1550 | No | ||
| 17 | WT1 | 1425995_s_at 1443221_at | 8709 | 0.619 | -0.1528 | No | ||
| 18 | TLX1 | 1450526_at | 8947 | 0.593 | -0.1590 | No | ||
| 19 | RXRA | 1425762_a_at 1430497_at 1454773_at | 9070 | 0.578 | -0.1601 | No | ||
| 20 | PPARA | 1449051_at | 9347 | 0.545 | -0.1684 | No | ||
| 21 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 9797 | 0.494 | -0.1851 | No | ||
| 22 | LMO1 | 1418478_at | 9907 | 0.480 | -0.1863 | No | ||
| 23 | ETV1 | 1422607_at 1445314_at 1450684_at | 10374 | 0.431 | -0.2043 | No | ||
| 24 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 10629 | 0.405 | -0.2127 | No | ||
| 25 | MYOD1 | 1418420_at 1447675_x_at | 10692 | 0.397 | -0.2125 | No | ||
| 26 | GLI3 | 1444043_at 1450525_at 1455154_at 1456067_at | 11110 | 0.350 | -0.2288 | No | ||
| 27 | MLLT7 | 1422149_at | 11962 | 0.255 | -0.2657 | No | ||
| 28 | FOXD1 | 1418876_at 1418877_at | 12767 | 0.167 | -0.3012 | No | ||
| 29 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 13156 | 0.123 | -0.3179 | No | ||
| 30 | JUN | 1417409_at 1448694_at | 13265 | 0.110 | -0.3220 | No | ||
| 31 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 13474 | 0.085 | -0.3309 | No | ||
| 32 | AHR | 1422631_at 1450695_at | 13740 | 0.047 | -0.3426 | No | ||
| 33 | FOSL2 | 1422931_at | 15047 | -0.146 | -0.4012 | No | ||
| 34 | PBX1 | 1430183_at 1440037_at 1440954_at 1442846_at 1449542_at 1458853_at | 15500 | -0.220 | -0.4201 | No | ||
| 35 | DPF2 | 1416534_at 1429903_at | 16242 | -0.380 | -0.4510 | No | ||
| 36 | IRF4 | 1421173_at 1421174_at | 16277 | -0.392 | -0.4495 | No | ||
| 37 | REL | 1420710_at | 16635 | -0.480 | -0.4621 | No | ||
| 38 | PAX7 | 1444596_at 1452510_at | 18346 | -1.097 | -0.5317 | No | ||
| 39 | NFKB2 | 1425902_a_at 1429128_x_at | 18571 | -1.202 | -0.5326 | Yes | ||
| 40 | ETS2 | 1416268_at 1437809_x_at 1447685_x_at | 18581 | -1.205 | -0.5236 | Yes | ||
| 41 | STAT5B | 1422102_a_at 1422103_a_at | 18936 | -1.397 | -0.5289 | Yes | ||
| 42 | SRF | 1418255_s_at 1418256_at | 19149 | -1.533 | -0.5266 | Yes | ||
| 43 | AATF | 1432394_a_at 1439676_at 1441722_at 1447459_at | 19167 | -1.543 | -0.5154 | Yes | ||
| 44 | STAT5A | 1421469_a_at 1450259_a_at | 19892 | -2.079 | -0.5322 | Yes | ||
| 45 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 20041 | -2.216 | -0.5217 | Yes | ||
| 46 | STAT6 | 1421708_a_at 1426353_at | 20465 | -2.672 | -0.5202 | Yes | ||
| 47 | TCF7L2 | 1425229_a_at 1426639_a_at 1429427_s_at 1429428_at 1441756_at 1446452_at 1447071_at | 20624 | -2.910 | -0.5047 | Yes | ||
| 48 | RELA | 1419536_a_at | 20799 | -3.184 | -0.4879 | Yes | ||
| 49 | PML | 1448757_at 1456103_at 1459137_at | 20925 | -3.435 | -0.4668 | Yes | ||
| 50 | PBX2 | 1418893_at 1418894_s_at 1449261_at | 20938 | -3.463 | -0.4403 | Yes | ||
| 51 | FOS | 1423100_at | 20955 | -3.496 | -0.4138 | Yes | ||
| 52 | SMARCA4 | 1426804_at 1426805_at | 21310 | -4.373 | -0.3959 | Yes | ||
| 53 | RELB | 1417856_at | 21351 | -4.570 | -0.3620 | Yes | ||
| 54 | JUNB | 1415899_at | 21503 | -5.184 | -0.3285 | Yes | ||
| 55 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 21749 | -7.027 | -0.2849 | Yes | ||
| 56 | ETV3 | 1418635_at 1418636_at 1445997_at 1456358_at | 21760 | -7.093 | -0.2301 | Yes | ||
| 57 | FLI1 | 1422024_at 1433512_at 1441584_at | 21767 | -7.169 | -0.1744 | Yes | ||
| 58 | BCL6 | 1421818_at 1450381_a_at | 21780 | -7.247 | -0.1184 | Yes | ||
| 59 | EGR1 | 1417065_at | 21860 | -7.832 | -0.0610 | Yes | ||
| 60 | EGR3 | 1421486_at 1436329_at | 21913 | -8.258 | 0.0011 | Yes |