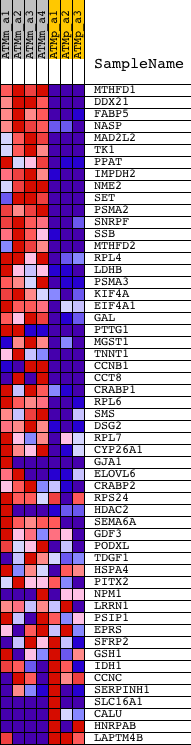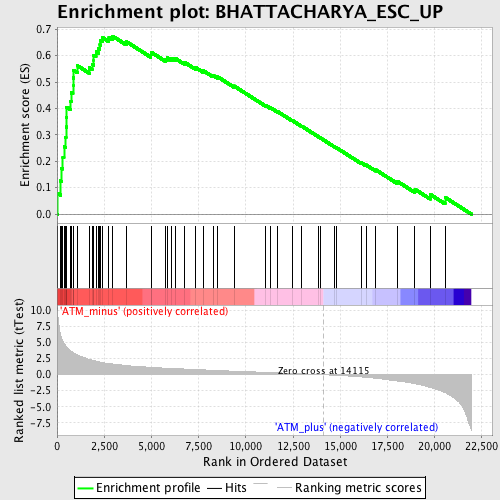
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | BHATTACHARYA_ESC_UP |
| Enrichment Score (ES) | 0.67368406 |
| Normalized Enrichment Score (NES) | 2.2369006 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MTHFD1 | 1415916_a_at 1415917_at 1436704_x_at | 30 | 9.067 | 0.0783 | Yes | ||
| 2 | DDX21 | 1448270_at 1448271_a_at | 178 | 6.224 | 0.1263 | Yes | ||
| 3 | FABP5 | 1416022_at | 228 | 5.702 | 0.1742 | Yes | ||
| 4 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 307 | 5.145 | 0.2158 | Yes | ||
| 5 | MAD2L2 | 1460348_at | 365 | 4.888 | 0.2562 | Yes | ||
| 6 | TK1 | 1416258_at | 458 | 4.505 | 0.2916 | Yes | ||
| 7 | PPAT | 1428543_at 1441770_at 1452831_s_at | 473 | 4.443 | 0.3300 | Yes | ||
| 8 | IMPDH2 | 1415851_a_at 1415852_at | 494 | 4.374 | 0.3675 | Yes | ||
| 9 | NME2 | 1448808_a_at | 507 | 4.339 | 0.4051 | Yes | ||
| 10 | SET | 1426853_at 1426854_a_at | 724 | 3.721 | 0.4279 | Yes | ||
| 11 | PSMA2 | 1448206_at | 744 | 3.668 | 0.4593 | Yes | ||
| 12 | SNRPF | 1428672_at | 842 | 3.476 | 0.4854 | Yes | ||
| 13 | SSB | 1416421_a_at 1416422_a_at 1416423_x_at 1453928_a_at | 849 | 3.465 | 0.5156 | Yes | ||
| 14 | MTHFD2 | 1419253_at 1419254_at | 876 | 3.397 | 0.5443 | Yes | ||
| 15 | RPL4 | 1425183_a_at | 1088 | 3.040 | 0.5614 | Yes | ||
| 16 | LDHB | 1416183_a_at 1434499_a_at 1442618_at 1448237_x_at 1455235_x_at | 1713 | 2.375 | 0.5537 | Yes | ||
| 17 | PSMA3 | 1448442_a_at | 1853 | 2.256 | 0.5672 | Yes | ||
| 18 | KIF4A | 1450692_at | 1938 | 2.190 | 0.5826 | Yes | ||
| 19 | EIF4A1 | 1427058_at 1430980_a_at 1434985_a_at | 1943 | 2.186 | 0.6016 | Yes | ||
| 20 | GAL | 1460668_at | 2061 | 2.103 | 0.6147 | Yes | ||
| 21 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 2201 | 2.007 | 0.6260 | Yes | ||
| 22 | MGST1 | 1415897_a_at 1415898_at | 2268 | 1.971 | 0.6403 | Yes | ||
| 23 | TNNT1 | 1419606_a_at | 2284 | 1.960 | 0.6569 | Yes | ||
| 24 | CCNB1 | 1419943_s_at 1419944_at | 2402 | 1.885 | 0.6681 | Yes | ||
| 25 | CCT8 | 1415785_a_at 1436973_at | 2745 | 1.733 | 0.6677 | Yes | ||
| 26 | CRABP1 | 1448326_a_at | 2936 | 1.667 | 0.6737 | Yes | ||
| 27 | RPL6 | 1416546_a_at | 3664 | 1.418 | 0.6529 | No | ||
| 28 | SMS | 1428699_at 1434190_at | 4972 | 1.138 | 0.6032 | No | ||
| 29 | DSG2 | 1425619_s_at 1426153_a_at 1439476_at 1449740_s_at 1449741_at 1460380_at | 4990 | 1.134 | 0.6124 | No | ||
| 30 | RPL7 | 1415979_x_at 1426162_a_at 1426163_x_at 1427452_at | 5751 | 1.015 | 0.5866 | No | ||
| 31 | CYP26A1 | 1419430_at | 5831 | 1.005 | 0.5918 | No | ||
| 32 | GJA1 | 1415800_at 1415801_at 1437992_x_at 1438650_x_at 1438945_x_at 1438973_x_at | 6078 | 0.976 | 0.5891 | No | ||
| 33 | ELOVL6 | 1417403_at 1417404_at 1445062_at 1445578_at 1445641_at | 6282 | 0.944 | 0.5882 | No | ||
| 34 | CRABP2 | 1451191_at | 6739 | 0.880 | 0.5751 | No | ||
| 35 | RPS24 | 1436064_x_at 1436500_at 1453362_x_at 1455195_at 1456628_x_at | 7333 | 0.795 | 0.5549 | No | ||
| 36 | HDAC2 | 1439704_at 1445684_s_at 1449080_at | 7741 | 0.744 | 0.5429 | No | ||
| 37 | SEMA6A | 1421414_a_at 1425903_at 1436458_at | 8262 | 0.675 | 0.5250 | No | ||
| 38 | GDF3 | 1449288_at 1460057_at | 8509 | 0.645 | 0.5195 | No | ||
| 39 | PODXL | 1417396_at 1448688_at | 9369 | 0.543 | 0.4850 | No | ||
| 40 | TDGF1 | 1450989_at | 11059 | 0.355 | 0.4109 | No | ||
| 41 | HSPA4 | 1416146_at 1416147_at 1440575_at | 11275 | 0.331 | 0.4040 | No | ||
| 42 | PITX2 | 1424797_a_at 1450482_a_at | 11687 | 0.286 | 0.3877 | No | ||
| 43 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 12454 | 0.203 | 0.3545 | No | ||
| 44 | LRRN1 | 1416053_at 1432715_at 1432716_at | 12957 | 0.147 | 0.3329 | No | ||
| 45 | PSIP1 | 1417166_at 1442148_at 1460403_at | 13844 | 0.035 | 0.2927 | No | ||
| 46 | EPRS | 1430370_at | 13969 | 0.019 | 0.2872 | No | ||
| 47 | SFRP2 | 1448201_at | 14687 | -0.088 | 0.2552 | No | ||
| 48 | GSH1 | 1450002_at | 14786 | -0.102 | 0.2516 | No | ||
| 49 | IDH1 | 1419821_s_at 1422433_s_at | 16095 | -0.346 | 0.1949 | No | ||
| 50 | CCNC | 1417861_at 1428570_at 1454144_a_at 1454336_at | 16367 | -0.412 | 0.1861 | No | ||
| 51 | SERPINH1 | 1450843_a_at 1456733_x_at | 16871 | -0.551 | 0.1679 | No | ||
| 52 | SLC16A1 | 1415802_at 1446274_at | 18010 | -0.986 | 0.1246 | No | ||
| 53 | CALU | 1415870_at 1441477_at | 18954 | -1.410 | 0.0939 | No | ||
| 54 | HNRPAB | 1415914_at 1426114_at 1431349_at 1448144_at 1453849_s_at 1455855_x_at 1459880_at | 19761 | -1.999 | 0.0746 | No | ||
| 55 | LAPTM4B | 1416148_at 1436915_x_at 1438365_x_at 1459215_at | 20549 | -2.812 | 0.0634 | No |