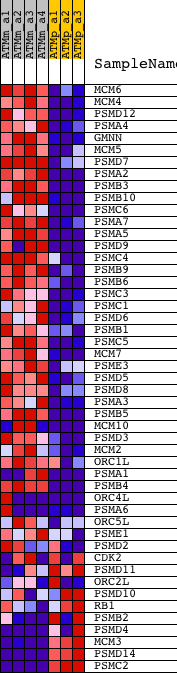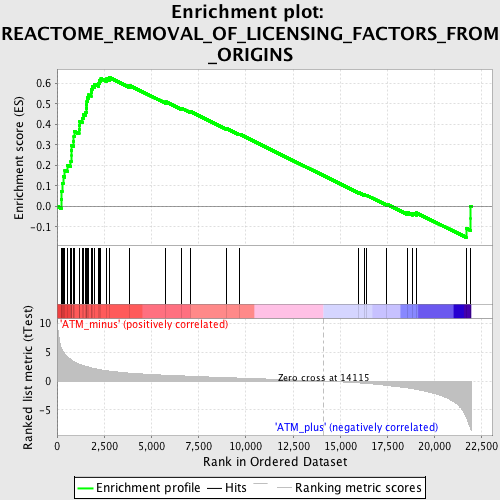
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_REMOVAL_OF_LICENSING_FACTORS_FROM_ORIGINS |
| Enrichment Score (ES) | 0.6289677 |
| Normalized Enrichment Score (NES) | 2.0856948 |
| Nominal p-value | 0.0 |
| FDR q-value | 3.5734338E-4 |
| FWER p-Value | 0.0060 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 213 | 5.833 | 0.0328 | Yes | ||
| 2 | MCM4 | 1416214_at 1436708_x_at | 225 | 5.728 | 0.0742 | Yes | ||
| 3 | PSMD12 | 1448492_a_at | 264 | 5.409 | 0.1119 | Yes | ||
| 4 | PSMA4 | 1460339_at | 318 | 5.099 | 0.1467 | Yes | ||
| 5 | GMNN | 1417506_at | 412 | 4.710 | 0.1768 | Yes | ||
| 6 | MCM5 | 1415945_at 1436808_x_at | 560 | 4.167 | 0.2005 | Yes | ||
| 7 | PSMD7 | 1432820_at 1451056_at 1454368_at | 729 | 3.702 | 0.2199 | Yes | ||
| 8 | PSMA2 | 1448206_at | 744 | 3.668 | 0.2460 | Yes | ||
| 9 | PSMB3 | 1417052_at | 747 | 3.666 | 0.2727 | Yes | ||
| 10 | PSMB10 | 1448632_at | 773 | 3.615 | 0.2979 | Yes | ||
| 11 | PSMC6 | 1417771_a_at | 882 | 3.386 | 0.3177 | Yes | ||
| 12 | PSMA7 | 1423567_a_at 1423568_at | 891 | 3.367 | 0.3419 | Yes | ||
| 13 | PSMA5 | 1424681_a_at 1434356_a_at | 919 | 3.308 | 0.3648 | Yes | ||
| 14 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 1161 | 2.949 | 0.3753 | Yes | ||
| 15 | PSMC4 | 1416290_a_at 1416291_at | 1172 | 2.934 | 0.3963 | Yes | ||
| 16 | PSMB9 | 1450696_at | 1201 | 2.903 | 0.4162 | Yes | ||
| 17 | PSMB6 | 1448822_at | 1328 | 2.765 | 0.4306 | Yes | ||
| 18 | PSMC3 | 1416282_at | 1398 | 2.695 | 0.4472 | Yes | ||
| 19 | PSMC1 | 1416005_at | 1516 | 2.575 | 0.4606 | Yes | ||
| 20 | PSMD6 | 1423696_a_at 1423697_at | 1546 | 2.546 | 0.4779 | Yes | ||
| 21 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 1566 | 2.522 | 0.4954 | Yes | ||
| 22 | PSMC5 | 1415740_at | 1578 | 2.503 | 0.5132 | Yes | ||
| 23 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 1610 | 2.476 | 0.5298 | Yes | ||
| 24 | PSME3 | 1418078_at 1418079_at 1438509_at | 1682 | 2.402 | 0.5441 | Yes | ||
| 25 | PSMD5 | 1423234_at | 1799 | 2.309 | 0.5557 | Yes | ||
| 26 | PSMD8 | 1423296_at | 1817 | 2.287 | 0.5716 | Yes | ||
| 27 | PSMA3 | 1448442_a_at | 1853 | 2.256 | 0.5865 | Yes | ||
| 28 | PSMB5 | 1415676_a_at | 1978 | 2.164 | 0.5966 | Yes | ||
| 29 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 2208 | 2.002 | 0.6007 | Yes | ||
| 30 | PSMD3 | 1448479_at | 2249 | 1.983 | 0.6134 | Yes | ||
| 31 | MCM2 | 1434079_s_at 1448777_at | 2322 | 1.939 | 0.6243 | Yes | ||
| 32 | ORC1L | 1422663_at 1443172_at | 2636 | 1.774 | 0.6229 | Yes | ||
| 33 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 2780 | 1.726 | 0.6290 | Yes | ||
| 34 | PSMB4 | 1438984_x_at 1451205_at | 3848 | 1.368 | 0.5902 | No | ||
| 35 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 5765 | 1.014 | 0.5101 | No | ||
| 36 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 6574 | 0.902 | 0.4797 | No | ||
| 37 | ORC5L | 1415830_at 1441601_at | 7063 | 0.831 | 0.4635 | No | ||
| 38 | PSME1 | 1417056_at | 8994 | 0.587 | 0.3796 | No | ||
| 39 | PSMD2 | 1415831_at | 9652 | 0.509 | 0.3533 | No | ||
| 40 | CDK2 | 1416873_a_at 1447617_at | 15987 | -0.320 | 0.0663 | No | ||
| 41 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 16256 | -0.383 | 0.0568 | No | ||
| 42 | ORC2L | 1418225_at 1418226_at 1418227_at | 16379 | -0.415 | 0.0543 | No | ||
| 43 | PSMD10 | 1436559_a_at | 17470 | -0.766 | 0.0101 | No | ||
| 44 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 18569 | -1.202 | -0.0313 | No | ||
| 45 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18820 | -1.334 | -0.0330 | No | ||
| 46 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 19036 | -1.455 | -0.0322 | No | ||
| 47 | MCM3 | 1420029_at 1426653_at | 21664 | -6.234 | -0.1067 | No | ||
| 48 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 21905 | -8.126 | -0.0584 | No | ||
| 49 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21908 | -8.187 | 0.0013 | No |