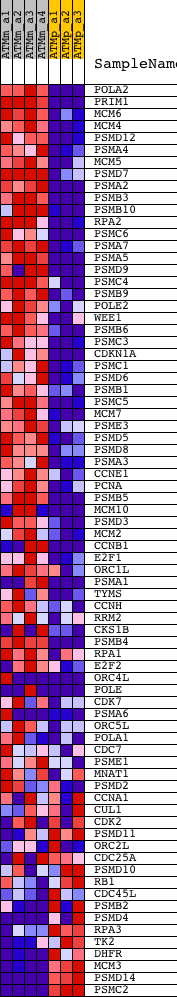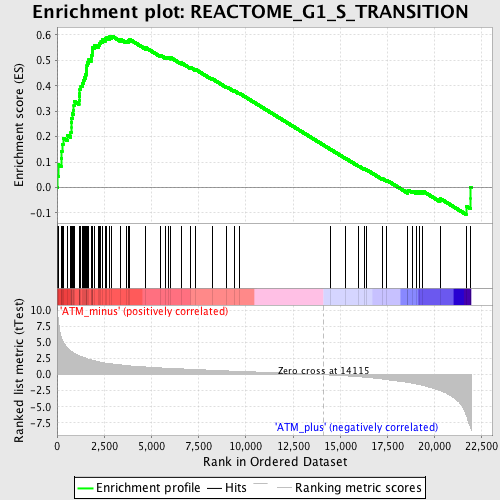
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_G1_S_TRANSITION |
| Enrichment Score (ES) | 0.59548813 |
| Normalized Enrichment Score (NES) | 2.1007533 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.3702034E-4 |
| FWER p-Value | 0.0020 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLA2 | 1425794_at 1448369_at 1456713_at | 39 | 8.968 | 0.0454 | Yes | ||
| 2 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 62 | 8.664 | 0.0900 | Yes | ||
| 3 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 213 | 5.833 | 0.1138 | Yes | ||
| 4 | MCM4 | 1416214_at 1436708_x_at | 225 | 5.728 | 0.1435 | Yes | ||
| 5 | PSMD12 | 1448492_a_at | 264 | 5.409 | 0.1702 | Yes | ||
| 6 | PSMA4 | 1460339_at | 318 | 5.099 | 0.1946 | Yes | ||
| 7 | MCM5 | 1415945_at 1436808_x_at | 560 | 4.167 | 0.2055 | Yes | ||
| 8 | PSMD7 | 1432820_at 1451056_at 1454368_at | 729 | 3.702 | 0.2173 | Yes | ||
| 9 | PSMA2 | 1448206_at | 744 | 3.668 | 0.2359 | Yes | ||
| 10 | PSMB3 | 1417052_at | 747 | 3.666 | 0.2551 | Yes | ||
| 11 | PSMB10 | 1448632_at | 773 | 3.615 | 0.2730 | Yes | ||
| 12 | RPA2 | 1416433_at 1454011_a_at | 818 | 3.519 | 0.2895 | Yes | ||
| 13 | PSMC6 | 1417771_a_at | 882 | 3.386 | 0.3045 | Yes | ||
| 14 | PSMA7 | 1423567_a_at 1423568_at | 891 | 3.367 | 0.3218 | Yes | ||
| 15 | PSMA5 | 1424681_a_at 1434356_a_at | 919 | 3.308 | 0.3380 | Yes | ||
| 16 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 1161 | 2.949 | 0.3425 | Yes | ||
| 17 | PSMC4 | 1416290_a_at 1416291_at | 1172 | 2.934 | 0.3574 | Yes | ||
| 18 | PSMB9 | 1450696_at | 1201 | 2.903 | 0.3714 | Yes | ||
| 19 | POLE2 | 1427094_at 1431440_at | 1207 | 2.896 | 0.3865 | Yes | ||
| 20 | WEE1 | 1416773_at 1416774_at | 1261 | 2.836 | 0.3990 | Yes | ||
| 21 | PSMB6 | 1448822_at | 1328 | 2.765 | 0.4105 | Yes | ||
| 22 | PSMC3 | 1416282_at | 1398 | 2.695 | 0.4215 | Yes | ||
| 23 | CDKN1A | 1421679_a_at 1424638_at | 1430 | 2.662 | 0.4341 | Yes | ||
| 24 | PSMC1 | 1416005_at | 1516 | 2.575 | 0.4438 | Yes | ||
| 25 | PSMD6 | 1423696_a_at 1423697_at | 1546 | 2.546 | 0.4558 | Yes | ||
| 26 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 1566 | 2.522 | 0.4682 | Yes | ||
| 27 | PSMC5 | 1415740_at | 1578 | 2.503 | 0.4809 | Yes | ||
| 28 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 1610 | 2.476 | 0.4925 | Yes | ||
| 29 | PSME3 | 1418078_at 1418079_at 1438509_at | 1682 | 2.402 | 0.5019 | Yes | ||
| 30 | PSMD5 | 1423234_at | 1799 | 2.309 | 0.5087 | Yes | ||
| 31 | PSMD8 | 1423296_at | 1817 | 2.287 | 0.5200 | Yes | ||
| 32 | PSMA3 | 1448442_a_at | 1853 | 2.256 | 0.5303 | Yes | ||
| 33 | CCNE1 | 1416492_at 1441910_x_at | 1864 | 2.245 | 0.5416 | Yes | ||
| 34 | PCNA | 1417947_at | 1885 | 2.232 | 0.5524 | Yes | ||
| 35 | PSMB5 | 1415676_a_at | 1978 | 2.164 | 0.5596 | Yes | ||
| 36 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 2208 | 2.002 | 0.5597 | Yes | ||
| 37 | PSMD3 | 1448479_at | 2249 | 1.983 | 0.5683 | Yes | ||
| 38 | MCM2 | 1434079_s_at 1448777_at | 2322 | 1.939 | 0.5752 | Yes | ||
| 39 | CCNB1 | 1419943_s_at 1419944_at | 2402 | 1.885 | 0.5815 | Yes | ||
| 40 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 2542 | 1.816 | 0.5847 | Yes | ||
| 41 | ORC1L | 1422663_at 1443172_at | 2636 | 1.774 | 0.5898 | Yes | ||
| 42 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 2780 | 1.726 | 0.5923 | Yes | ||
| 43 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 2905 | 1.680 | 0.5955 | Yes | ||
| 44 | CCNH | 1418585_at | 3379 | 1.514 | 0.5818 | No | ||
| 45 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 3670 | 1.417 | 0.5760 | No | ||
| 46 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 3781 | 1.387 | 0.5783 | No | ||
| 47 | PSMB4 | 1438984_x_at 1451205_at | 3848 | 1.368 | 0.5825 | No | ||
| 48 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 4687 | 1.195 | 0.5504 | No | ||
| 49 | E2F2 | 1455790_at 1458574_at | 5492 | 1.050 | 0.5192 | No | ||
| 50 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 5765 | 1.014 | 0.5120 | No | ||
| 51 | POLE | 1441854_at 1448650_a_at | 5885 | 1.000 | 0.5119 | No | ||
| 52 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 6028 | 0.983 | 0.5105 | No | ||
| 53 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 6574 | 0.902 | 0.4904 | No | ||
| 54 | ORC5L | 1415830_at 1441601_at | 7063 | 0.831 | 0.4724 | No | ||
| 55 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 7331 | 0.795 | 0.4644 | No | ||
| 56 | CDC7 | 1426002_a_at 1426021_a_at | 8225 | 0.679 | 0.4271 | No | ||
| 57 | PSME1 | 1417056_at | 8994 | 0.587 | 0.3951 | No | ||
| 58 | MNAT1 | 1432177_a_at | 9371 | 0.543 | 0.3807 | No | ||
| 59 | PSMD2 | 1415831_at | 9652 | 0.509 | 0.3706 | No | ||
| 60 | CCNA1 | 1449177_at | 14468 | -0.053 | 0.1506 | No | ||
| 61 | CUL1 | 1448174_at 1459516_at | 15253 | -0.177 | 0.1157 | No | ||
| 62 | CDK2 | 1416873_a_at 1447617_at | 15987 | -0.320 | 0.0838 | No | ||
| 63 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 16256 | -0.383 | 0.0736 | No | ||
| 64 | ORC2L | 1418225_at 1418226_at 1418227_at | 16379 | -0.415 | 0.0702 | No | ||
| 65 | CDC25A | 1417131_at 1417132_at 1445995_at | 17222 | -0.670 | 0.0352 | No | ||
| 66 | PSMD10 | 1436559_a_at | 17470 | -0.766 | 0.0279 | No | ||
| 67 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 18569 | -1.202 | -0.0160 | No | ||
| 68 | CDC45L | 1416575_at 1457838_at | 18583 | -1.205 | -0.0102 | No | ||
| 69 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18820 | -1.334 | -0.0140 | No | ||
| 70 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 19036 | -1.455 | -0.0162 | No | ||
| 71 | RPA3 | 1448938_at | 19181 | -1.555 | -0.0146 | No | ||
| 72 | TK2 | 1426100_a_at | 19373 | -1.685 | -0.0145 | No | ||
| 73 | DHFR | 1419172_at 1430750_at | 20280 | -2.447 | -0.0430 | No | ||
| 74 | MCM3 | 1420029_at 1426653_at | 21664 | -6.234 | -0.0735 | No | ||
| 75 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 21905 | -8.126 | -0.0417 | No | ||
| 76 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21908 | -8.187 | 0.0013 | No |