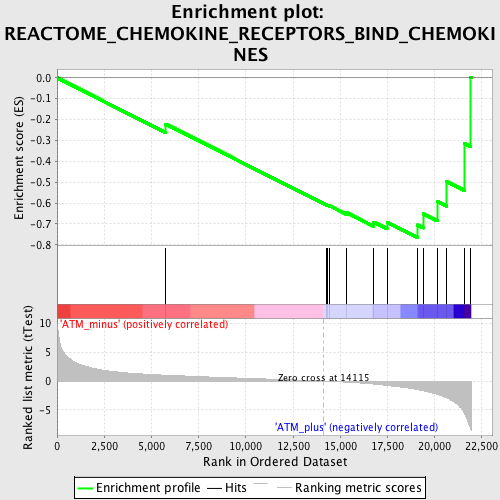
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES |
| Enrichment Score (ES) | -0.7646887 |
| Normalized Enrichment Score (NES) | -1.8734441 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.023004092 |
| FWER p-Value | 0.254 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 5754 | 1.015 | -0.2215 | No | ||
| 2 | IL8RB | 1421734_at | 14264 | -0.024 | -0.6086 | No | ||
| 3 | CCL20 | 1422029_at | 14330 | -0.033 | -0.6103 | No | ||
| 4 | CXCL11 | 1419697_at 1419698_at | 14439 | -0.049 | -0.6132 | No | ||
| 5 | XCR1 | 1422294_at | 15329 | -0.192 | -0.6460 | No | ||
| 6 | CCR3 | 1422957_at | 16774 | -0.523 | -0.6908 | No | ||
| 7 | CCR10 | 1421420_at | 17475 | -0.769 | -0.6917 | No | ||
| 8 | CXCR6 | 1422812_at 1425832_a_at | 19076 | -1.483 | -0.7049 | Yes | ||
| 9 | CCR6 | 1450357_a_at | 19395 | -1.700 | -0.6508 | Yes | ||
| 10 | XCL1 | 1419412_at | 20131 | -2.299 | -0.5916 | Yes | ||
| 11 | CCL25 | 1418777_at 1458277_at | 20636 | -2.929 | -0.4964 | Yes | ||
| 12 | CCL5 | 1418126_at | 21570 | -5.584 | -0.3136 | Yes | ||
| 13 | CCR7 | 1423466_at | 21909 | -8.188 | 0.0012 | Yes |

